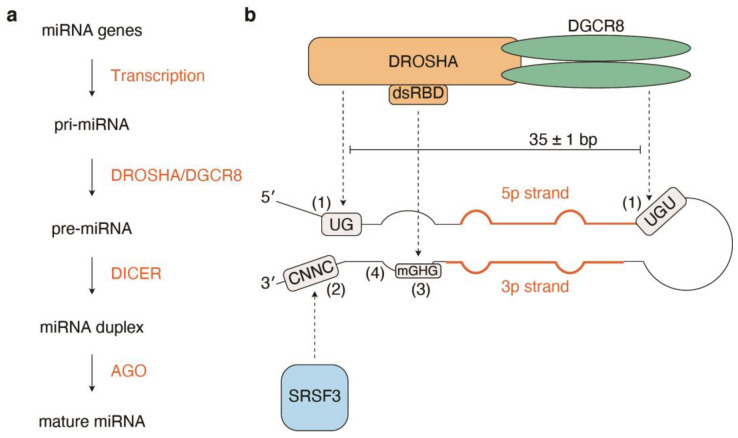
Figure 1.
Structural basis of efficient pri-miRNA processing. (a) Outline of the canonical miRNA biogenesis. (b) The structure of pri-miRNA and related molecules are demonstrated. The numbers in parentheses indicate the features of the miRNA precursor sequences contributing to efficient pri-miRNA processing. (1) UG and UGU motifs are recognized by DROSHA and DGCR8, respectively. Thus, the DROSHA–DGCR8 complex contributes to the correct measurement of the miRNA basal stem length. (2) A CNNF motif is bound by SRSF3. SRSF3s have multifaceted roles in miRNA processing by facilitating pri-miRNA processing efficiency, modulating alternative processing, and preventing 5p-nick processing and inverse processing. (3) An mGHG motif is bound by the dsRBD of DROSHA and helps DROSHA identify the miRNA cleavage site. (4) The large-sized asymmetric internal loops, i.e., large-sized mismatches, inhibit the cleavage of the 3p strand and therefore downregulate miRNA expression. Pri-miRNA, primary miRNA; mGHG, mismatched GHG; dsRBD, double-stranded RNA binding domain.