Fig. 6.
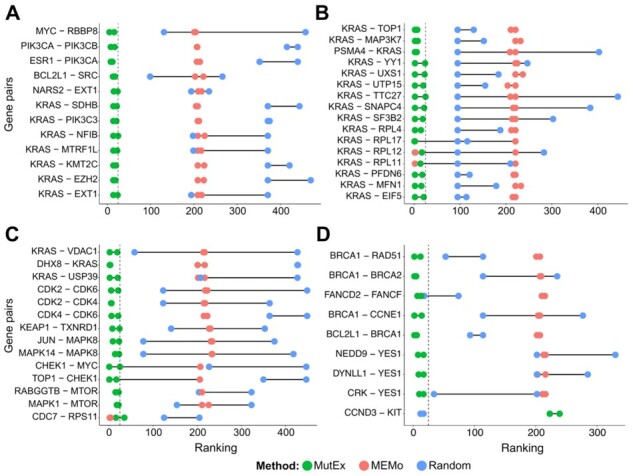
Comparison between MutExMatSorting, MEMo and random strategies in highlighting known human synthetic lethal pairs across different cancer types. Row-based ranking of known cancer-specific synthetic lethal (SL) pairs in breast carcinoma (A), colorectal carcinoma (B), lung adenocarcinoma (C) and ovarian carcinoma (D) upon MutExMatSorting, MEMo or random sorting of the binary dependency dataset derived from the Project Score. Only SL pairs with a confidence score > 0.4 were selected. For each cancer type, we subset the binary dataset considering the corresponding cancer cell lines and genes part of the SL pairs. We also added 200 core-fitness and 200 never-essential genes as negative controls (i.e. non-mutually exclusive). The figures show only the SL pairs found in the top 25 rows (dashed line) of at least one of the sorted matrices
