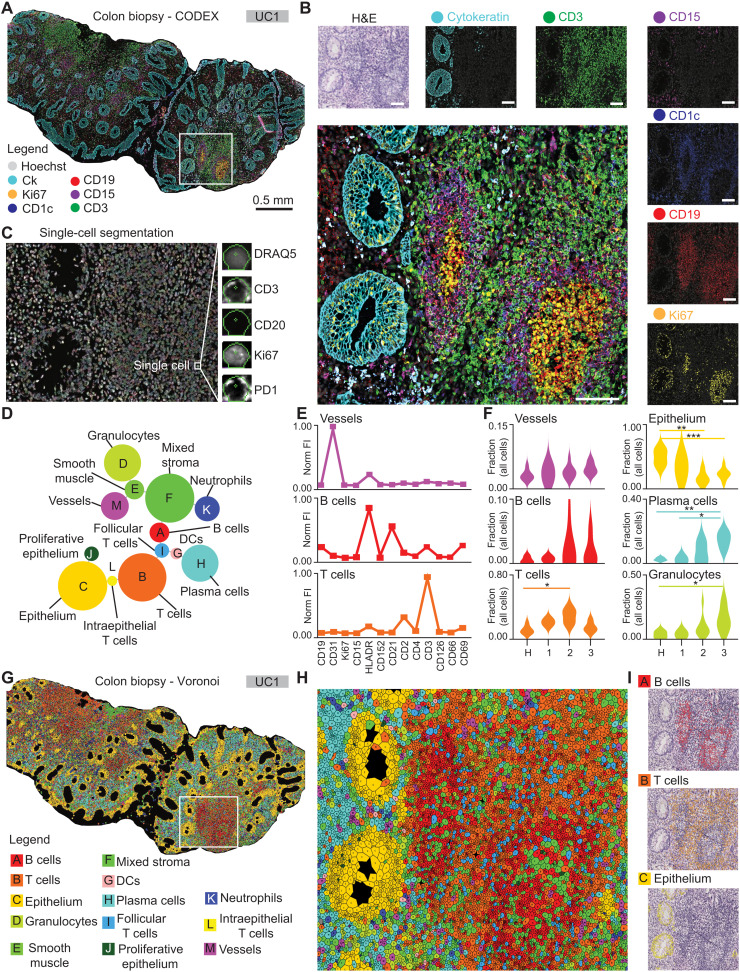
Fig. 1. A spatial atlas of the UC inflammatory microenvironment.
(A) CODEX image with Hoechst nuclear stain (gray), cytokeratin (cyan), Ki67 (yellow), CD1c (blue), CD19 (red), CD15 (purple), and CD3 (green) shown as a seven-color composite image selected from five markers stained on the same tissue section. (B) Zoomed-in view of the region inside the white box in (A) shown as a seven-color composite image (large panel) and as hematoxylin and eosin (H&E) and two-color images of Hoechst and each indicated marker individually (small panels). (C) Representative example of single-cell segmentation within the same box with DRAQ5 nuclear staining (gray) and the segmentation masks overlaid (colored). (D) Minimum spanning tree of the cell-type clusters identified in this study. Nodes represent clusters with sizes indicative of the number of cells across all samples, and distances between nodes indicate their relationships in high-dimensional marker space, not physical space. (E) Examples of marker expression profiles, in normalized fluorescent intensity (Norm FI) were extracted and used for the identification of cell types. (F) Single-cell clustering results and cell types across the entire cohort, evaluated at the patient level. H, healthy controls; 1 to 3, patients with UC with Mayo scores 1 to 3, respectively. Statistics: Student’s t test with Bonferroni correction for multiple comparisons (diseased versus healthy); *P < 0.05, **P < 0.01, and ***P < 0.001. (G and H) Voronoi representations of the identified cell types on the same tissue section as shown in (A) and (B). (G) Biopsy overview and legend of cell types. (H) Zoomed-in view of the region denoted in the white box in (G). (I) Example mapping of clustering results to H&E images used to validate cell type identity “A” (B cells, top), “B” (T cells, middle), and “C” (epithelium, bottom).