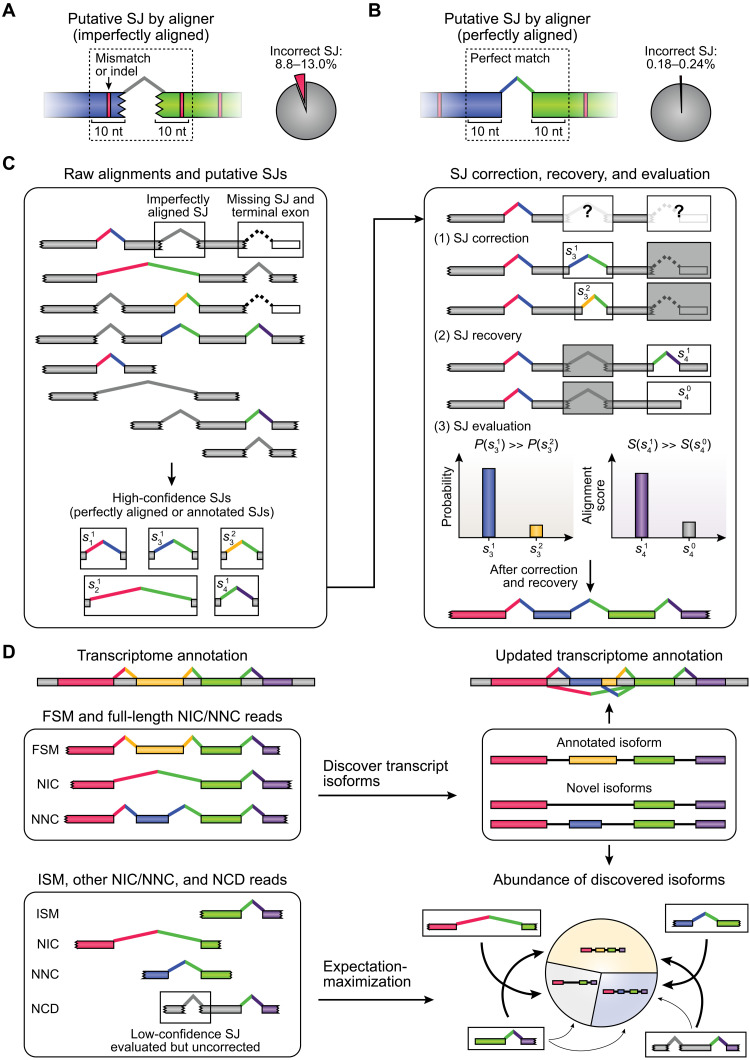
Fig. 1. Overview of ESPRESSO.
(A and B) Proportion of incorrect splice junctions (SJs) among (A) imperfectly aligned or (B) perfectly aligned putative SJs found in raw long-read-to-genome alignments of ONT 1D cDNA reads (n = 3) and direct RNA reads (n = 3) for Spike-In RNA Variants (SIRVs). Perfectly aligned putative SJs do not have any mismatches or indels within 10 nt of splice sites. (C) High-confidence SJs are identified from raw long-read-to-genome alignments based on whether they are present in the existing transcript catalog, or if they have canonical splice site dinucleotide motifs (GT/AG, GC/AG, or AT/AC) and are supported by at least two (by default) perfectly aligned reads. The resulting set of high-confidence SJs is used to correct, recover, and evaluate SJs found in individual long reads based on each read’s alignment and error profile. (D) First, reads are classified into the following categories on the basis of the annotation statuses of their corresponding SJs in the existing transcript catalog: full splice match (FSM), incomplete splice match (ISM), novel in catalog (NIC), novel not in catalog (NNC), or not completely determined (NCD). Second, FSM and full-length NIC/NNC reads are used to discover annotated and novel transcript isoforms, respectively. Third, all long reads (full-length and non-full-length) are matched to compatible transcript isoforms. Last, abundances of discovered isoforms are quantified using an expectation-maximization (EM) algorithm. Thickness of arrows drawn between reads and compatible transcript isoforms (bottom right) indicates probability of assigning reads to specific transcript isoforms.