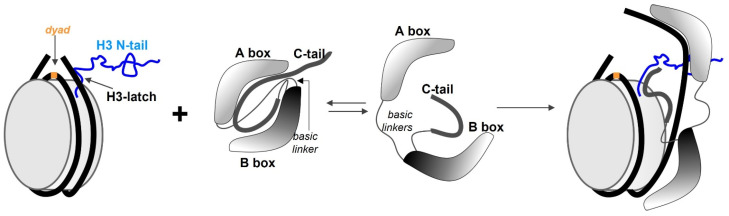
Figure 3.
The interaction of HMGB1 with the NCP. Schematic representation of nucleosomal DNA (black), a histone octamer (a grey cylinder), HMGB1 (A and B boxes, two basic linkers and the acidic C-tail) and the basic N-tail of H3. The dyad is highlighted in orange. In the absence of DNA, HMGB1 is in a closed conformation that is transformed by non-sequence-specific interaction with linker DNA at the entry/exit site of NCP. The complex is stabilised by the interaction between the acidic tail of HMGB1 and N- tail of H3; the interaction drives DNA unwrapping and NCP destabilisation.