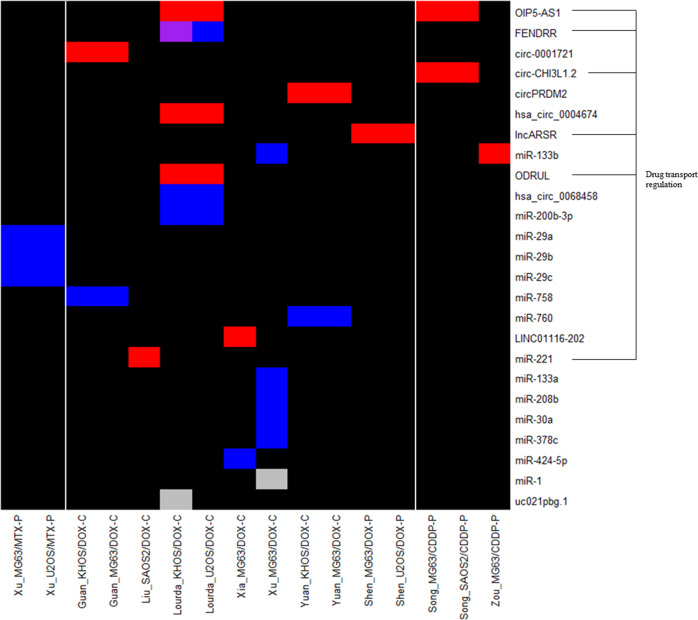
Fig. 3. Differential ncRNA expression in chemoresistant OS models.
ncRNA expression data for in vitro studies was extracted into a data table and entered into R to develop a summary heatmap of expression grouped by distinct model. ncRNAs are organised from most (top) to least (bottom) investigated across different models. Models are ordered by the MAP agent used for generation. (Blue) Downregulated, (Red) Upregulated, (Grey) Unchanged with respect to each models’ corresponding parental cell line, (Purple) Differing expression results within the same model, (Black) unknown or ambiguous result indicating an area of potential research interest. ncRNAs shown to be involved in the regulation of drug efflux proteins are highlighted due to their occurrence (number of studies = 15).