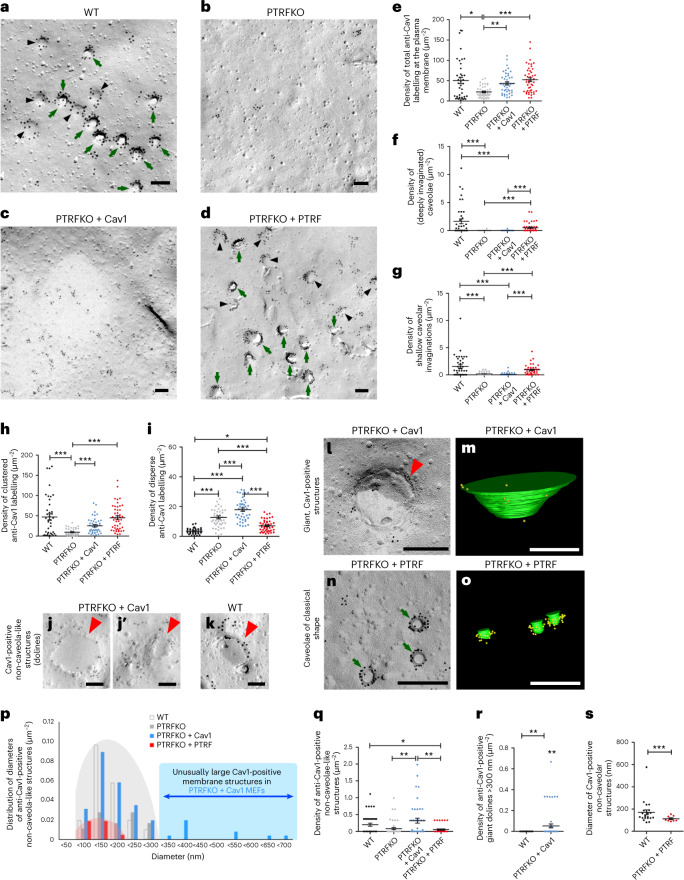
Fig. 5. PTRF genetic ablation leads to loss of caveolae and accumulation of non-caveolar Cav1 immunopositive structures.
a–d, Anti-Cav1 immunogold-labelled PMs of WT (a), PTRFKO (b), PTRFKO + Cav1 (c) and PTRFKO + PTRF MEFs (d). Caveolae (a, arrows) and anti-Cav1-positive shallow invaginations (a, arrowheads) were absent in b and c. Scale bars, 100 nm. e, Anti-Cav1 immunolabelling density across all four MEFs genotypes. f,g, Caveolae densities (f, deep; g, shallow). h,i, Densities of ‘clustered’ (h) and ‘disperse’ (i) anti-Cav1 immunogold labels. j,k, Cav1-immunopositive non-caveolar structures (red arrowheads) in PTRFKO + Cav1 MEFs (j and j′], morphologically distinct from caveolae, are also found (usually smaller) in WT cells (k). Scale bars, 100 nm. l–o, Electron tomograms (l and n) and 3D reconstructions (m and o) of a non-caveolar structure at the PM of PTRFKO + Cav1 cells (l and m), and ‘classical’ caveolae in PTRFKO + PTRF cells (n and o). Marks as in a, j, j′ and k, respectively. In m and o, gold particles: yellow; PM: green. Scale bars, 200 nm. p, Density distribution of non-caveolar anti-Cav1-positive structures in PRTFKO + PTRF and WT cells (grey shadowed area; diameters up to 299 nm); and in PTRFKO + Cav1 cells (blue shadowed area; diameters up to 300–699 nm). Red shadowing: PTRFKO + PTRF cells. q–s, Density analyses of all (q) and large (r, diameter ≥300 nm) non-caveolar Cav1-immunopositive structures; diameters in s. Plots: mean ± s.e.m.; WT control, n = 40 images per total ROI area 104.25 µm2; PTRFKO, n = 46 per 214.75 µm2; PTRFKO + Cav1, n = 44 per 258.4 µm2; PTRFKO + PTRF, n = 46 per 182.01 µm2 from three independent experiments (e–i, q and r). For non-caveolar structure characterization: WT, n = 22; PTRFKO, n = 18; PTRFKO + Cav1, n = 70 and PTRFKO + PTRF, n = 9 (p and s). The technique does not allow to determine how many cells are analysed, as only patches of cell membrane are observed. Statistical analyses: Kruskal–Wallis test with Dunn’s post-test (e, P values: *0.048; **0.001; ***<0.0001; f, P values: all ***<0.0001; g, P values: all ***<0.0001; h, P values: WT versus PTRFKO < 0.0001; PTRFKO versus PTRFKO + Cav1 0.008; PTRFKO versus PTRFKO + PTRF <0.0001), one-way ANOVA with Tukey post-test (i, P values: all ***<0.0001; *0.0136), and two-tailed Student’s t-test (r, P = 0.0044, s, P = 0.0002), respectively. Source numerical data are available in source data.