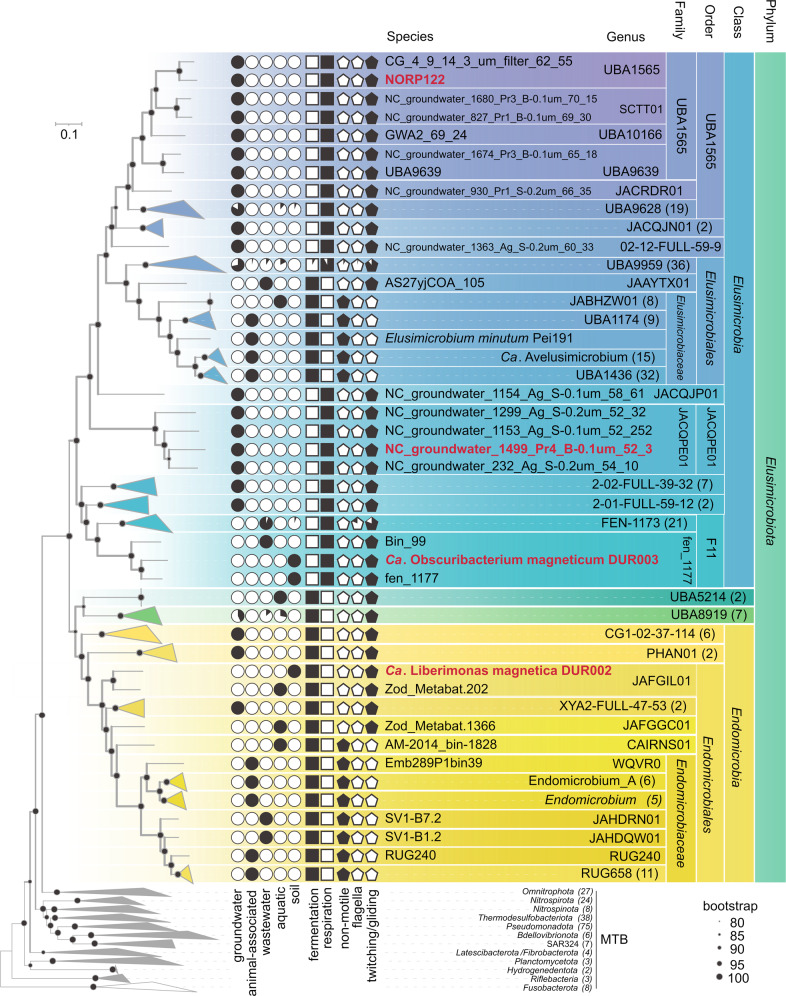
Fig. 3. Maximum-likelihood phylogenomic tree of all known Elusimicrobiota, all MTB genomes previously known or obtained in this work.
The tree was inferred from 120 concatenated bacterial single-copy marker proteins constructed with the evolutionary model LG + F + I + G4. Branch supports were obtained with 1000 ultrafast bootstraps. The scale bar represents amino acid substitutions per site. MTB genomes studied in this work are highlighted in red. Circles depict the habitat preferences of all studied genomes. Squares indicate the presence of fermentation-based or respiratory-based metabolism genes. Pentagons reflect the presence or absence of genes for a certain motility type.