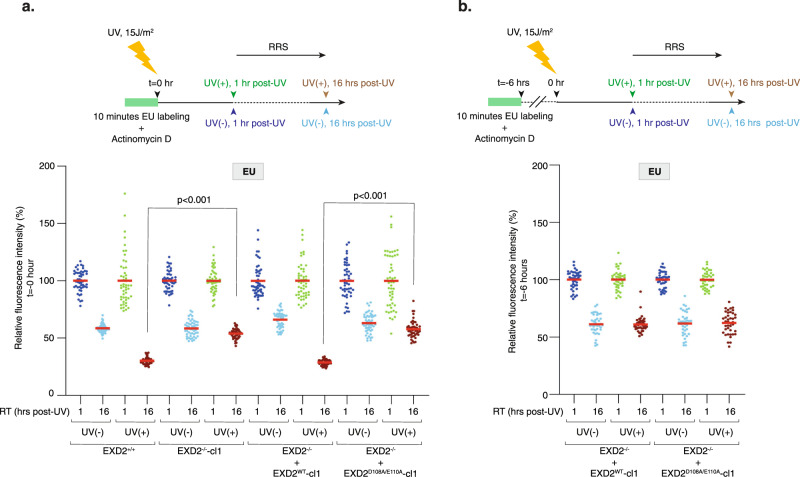
Fig. 4. EXD2 degrades mRNA under synthesis at the time of UV irradiation.
a Upper panel; Scheme of the EU pulse-chase method used to analyze the fate of mRNA being synthesized at the moment of UV irradiation. Cells were incubated for 30 min with Actinomycin D (0.05 μg/ml) to specifically inhibit RNAPI transcription and mRNAs were pulse-labeled with EU for 10 min prior to UV irradiation (15 J/m2). Cells were let to recover for 1 h or 16 h post-UV before fixation. Actinomycin D was maintained during the experiment. Lower panel; Cells were treated as indicated in the upper panel and EU signals were quantified using ImageJ and normalized to the value obtained at 1 h set to 100%. Values are reported on the graph (n = at least 50 cells). Red bars indicate mean integrated density. RT; recovery time. One-way ANOVA with post-hoc Tukey adjustment comparisons were used to determine the p-values. Source data are provided as a Source Data file. b Upper panel; Compare to panel a, UV irradiation (15 J/m2) was performed 6 h after EU labeling and cells were let to recover for either 1 h or 16 h post-UV before fixation. Actinomycin D was maintained during the experiment. Lower panel; Cells were treated as indicated in upper panel and EU signals were quantified using ImageJ. Values are reported on the graph (n = at least 50 cells). Red bars indicate mean integrated density. RT recovery time. One-way ANOVA with post-hoc Tukey adjustment comparisons were used to determine the p-values. No statistically significant differences were detected at 16 h post-UV between EXD2−/− + EXD2WT-cl1 and EXD2−/− + EXD2D108A/E110A-cl1 cells. Source data are provided as a Source Data file.