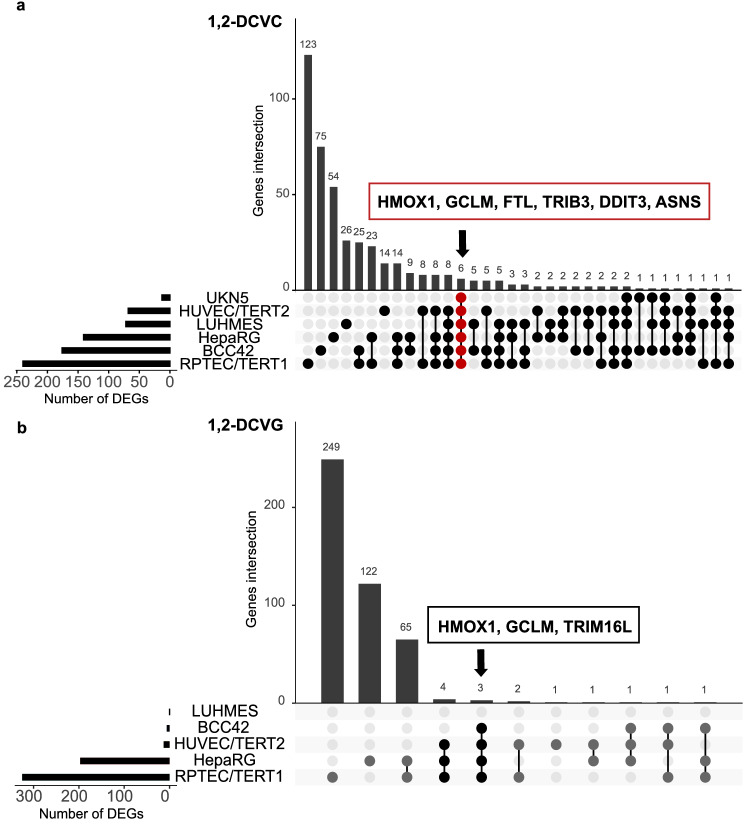
Fig. 5.
Intersection of upregulated DEGs for the top concentrations of 1,2-DCVC and 1,2-DCVG exposure across cell models. Number of DEGs: number of upregulated DEGs found for cell model (padj < 0.01, basemean > 35, log2FC > 0.58). Genes intersection: Number of DEGs found in common for depicted cell models (black or red dots) a 1,2-DCVC exposure. Six genes found commonly upregulated across the cell panel: Heme Oxygenase 1 (HMOX1), Glutamate-Cysteine Ligase (GCLM) and Ferritin Light Chain (FTL), DNA Damage Inducible Transcript 3 (DDIT3), Tribbles Pseudokinase 3 (TRIB3) and Asparagine Synthetase (Glutamine-Hydrolyzing) (ASNS). b 1,2-DCVG exposure. UKN5 were not tested for 1,2-DCVG exposure and therefore not included in the analysis. No DEGs were found after 1,2-DCVG exposure in LUHMES. RPTEC/TERT1, HUVEC/TERT2 and HepaRG demonstrated three genes commonly upregulated, HMOX1, GCLM and Tripartite Motif Containing 16 Like (TRIM16L)