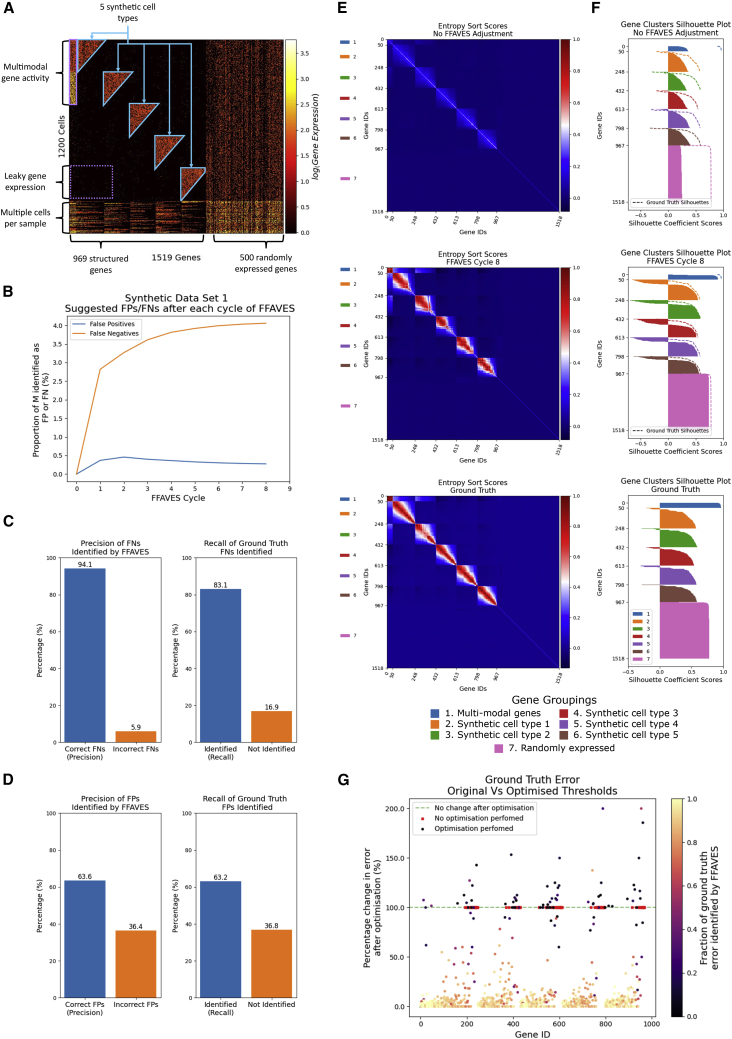
Figure 4.
FFAVES accurately identifies false negatives and false positives
(A) The synthetic scRNA-seq dataset.
(B) Convergence of FN/FP data points identified after each cycle of FFAVES.
(C and D) Precision and recall scores of FNs and FPs identified by FFAVES, respectively.
(E) Heatmaps of pairwise feature ESSs. Top: before identification of FNs and FPs by FFAVES. Middle: after application of FFAVES. Bottom: ground truth, i.e., synthetic data prior to introduction of FN dropouts.
(F) Silhouette scores of the seven main gene groups calculated from the respective ESSs in (E). Dashed lines outline the ground truth silhouette scores.
(G) Reduction in FN errors that were intentionally introduced by sub-optimal feature discretization.