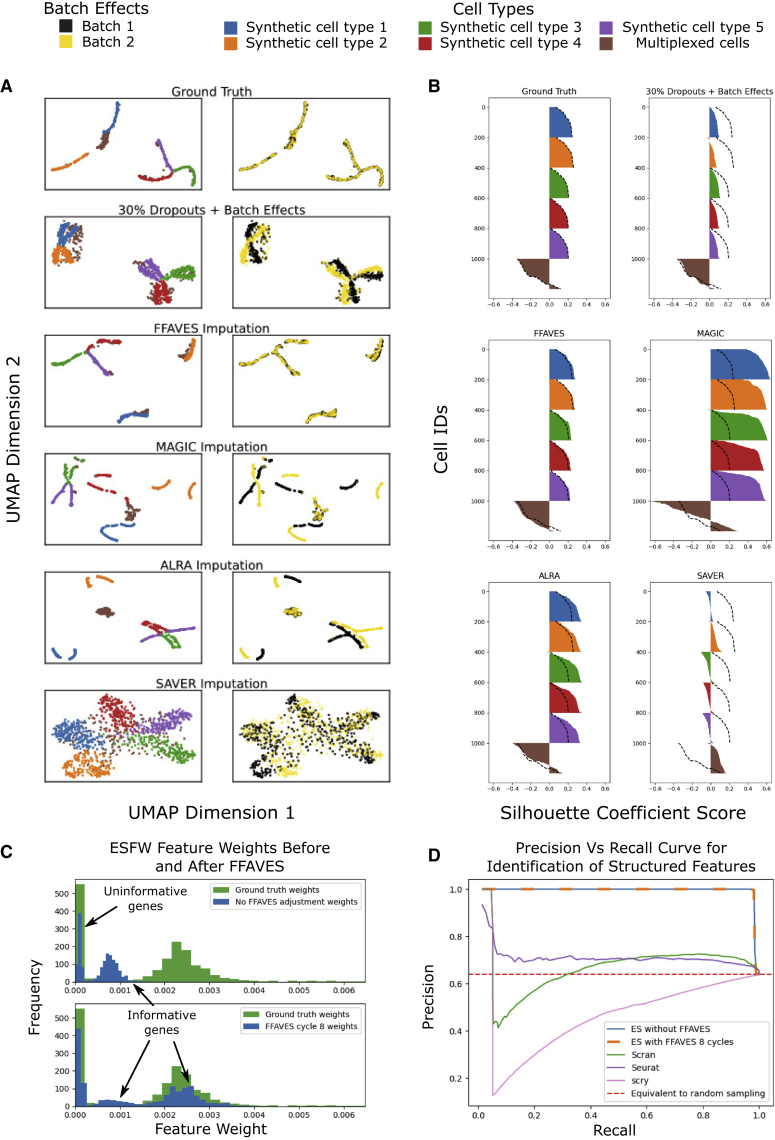
Figure 5.
Performance of FFAVES and ESFW against comparable software
(A) UMAPs of the synthetic dataset before and after imputation. The top two plots show the synthetic data before and after FN dropouts were introduced, with no imputation.
(B) Silhouette scores for each of the six main clusters of cells in the synthetic dataset. Black dashed lines mark the silhouette scores of the ground truth data prior to the introduction of FNs.
(C) Feature importance weights for all genes in the synthetic data according to ESFW. Top: feature weights estimated from the synthetic data with FNs introduced to the ground truth. Bottom: feature weights estimates after FFAVES has identified statistically significant divergent data points.
(D) Precision/recall curves for distinguishing structured and randomly expressed genes. Each line is generated from the ranked gene lists of the respective feature selection software.