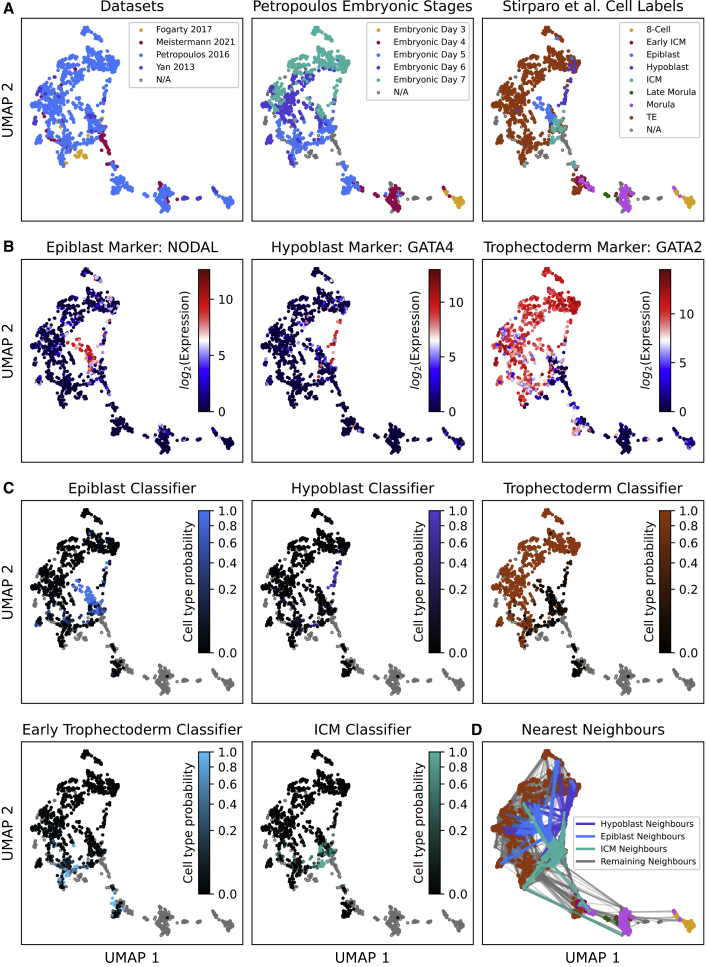
Figure 6.
Independent validation of the FFAVES + ESFW human pre-implantation embryo embedding
(A) The FFAVES + ESFW UMAP embedding overlaid with different label information: (left to right) the datasets that samples originate from, the time point labels from the Petropoulos dataset, and the cell type labels for the Petropoulos dataset that were independently assigned by Stirparo et al., (2018).
(B) Example epiblast, hypoblast, and trophectoderm marker expression. See Figure S8 for more examples.
(C) Predicted cell type probabilities of individual cells from a classifier trained on human pre-implantation embryo scRNA-seq data from the independent Yanagida et al., (2021) dataset. Gray samples are those that were not processed by the classifier to avoid confounding variables such as batch effects. See Figure S6B for the same analysis with Macaca classifiers.
(D) Nearest neighbor embedding where each cell is connected by lines to their 10 most similar samples according to gene expression. See Figure S6C for individual cell type nearest neighbor embeddings.