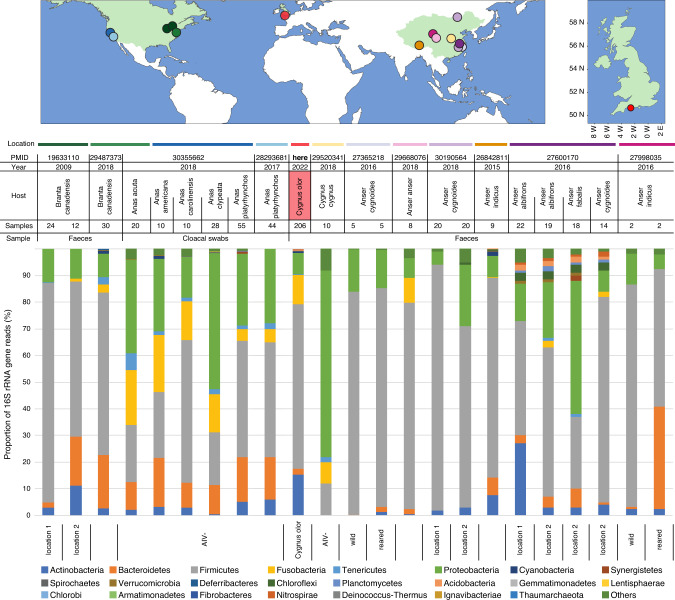
Fig. 1. Comparison of the composition of faecal prokaryotic communities of C. olor, at the phylum level, to the results obtained by other metagenomic studies conducted on wild Anatidae species.
The bibliographic search was conducted of PubMed on 16 September 2019 using the Adjutant package in R, using the search ‘(bird OR avian) AND (bact metagenomic OR 16S OR bact microbio)’ (1424 papers). We removed papers (i) that were not primary research articles, (ii) whose samples were not taken from living birds, (iii) that targeted one or few specific prokaryote taxa, and (iv) that did not target wild populations of Anatidae species. We consequently retained a total of 11 papers on Anatidae prokaryotic microbiomes (corresponding PMID are provided on top of the figure). Proportion of 16S rRNA gene reads at the phylum level was obtained by screening the results of these 11 published studies. The upper maps show the approximate location of sampling of each of these studies as indicated by corresponding coloured dots and horizontal bars.