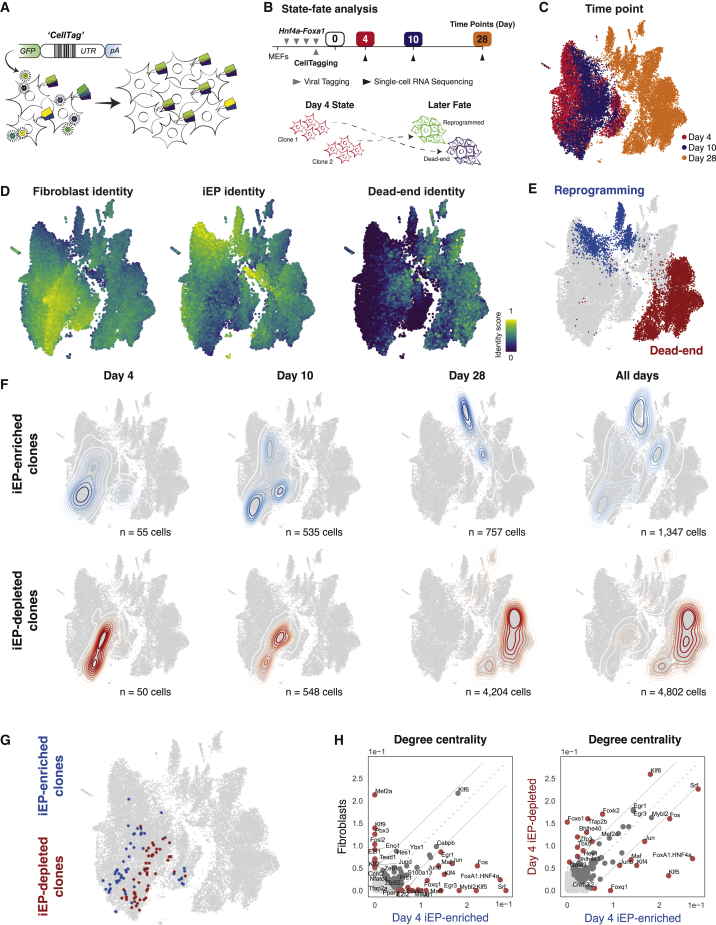
Figure 2.
Lineage tracing links early network state to reprogramming outcome
(A) Overview of CellTag-based clonal tracking. Cells are transduced with the random CellTag lentiviral library so that each cell expresses three to four CellTags, resulting in a unique, heritable barcode signature. CellTags are transcribed and captured during single-cell profiling, enabling clonally related cells to be tracked throughout an experiment.
(B) Experimental strategy to capture state-fate relationships. MEFs are transduced with Hnf4α-Foxa1 for 48 h, then transduced with CellTags. The end of this period is considered reprogramming day 0. Cells are expanded, and 25% of the population is profiled at day 4; this is termed the state population. The remaining cells are reseeded and profiled again on days 10 and 28 to capture reprogramming fate.
(C) Captured state-fate cells. Time point information projected onto the Uniform Manifold Approximation and Projection (UMAP) embedding. A total of 24,799 cells were sequenced: 8,440 on day 4, 4,836 on day 10, and 11,523 on day 28.
(D) Projection of fibroblast, iEP, and dead-end identity scores and (E) fate annotations onto the UMAP embedding.
(F) A randomized test identified day 4 state clones whose day 10 and 28 fate sisters were iEP-enriched or iEP-depleted. (Top) Kernel density estimation of iEP-enriched day 4 state clones and their day 10 and 28 fates, outlining the reprogramming trajectory (n = 1,347 cells). (Bottom) iEP-depleted state-fate cells outlining the dead-end trajectory (n = 4,802 cells).
(G) Projection of iEP-enriched and iEP-depleted clones onto the UMAP embedding.
(H) Comparison of degree centrality scores between native fibroblasts and day 4 reprogrammed-destined cells (left) and day 4 reprogrammed- and dead-end-destined cells (right).