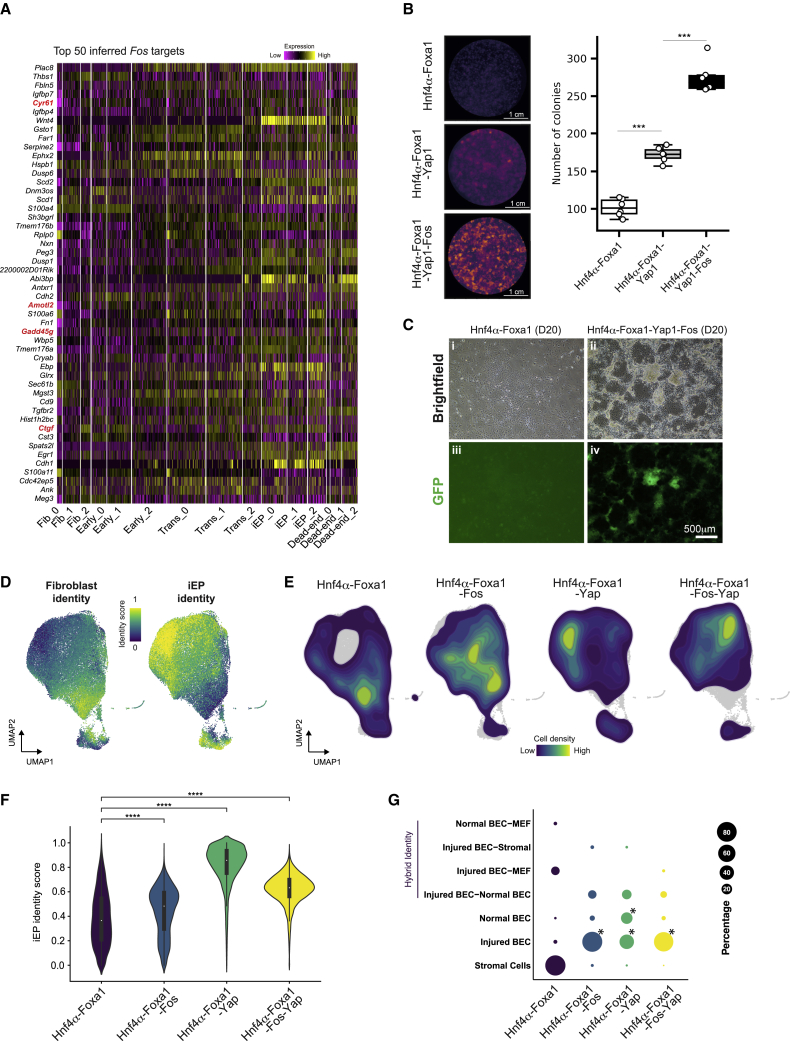
Figure 5.
Inferred Fos targets reveal a role for the Hippo signaling effector, Yap1, in reprogramming
(A) Heatmap of expression of the top 50 inferred Fos targets across reprogramming. Established YAP1 targets are highlighted in red.
(B) Colony-formation assay with the addition of Yap1 and Fos to Hnf4α-Foxa1. (Left) E-cadherin immunohistochemistry. (Right) Boxplot of colony numbers (n = 6 independent biological replicates; ∗∗∗p < 0.001, t-test, one sided).
(C) Brightfield and epifluorescence images of cells reprogrammed with Hnf4α-Foxa1 or Hnf4α-Foxa1-Fos-Yap1. Scale bar, 500 μm.
(D) scRNA-seq of cells reprogrammed with Hnf4α-Foxa1 (n = 7,414 cells), Hnf4α-Foxa1-Fos (n = 8,771 cells), Hnf4α-Foxa1-Yap1 (n = 8,549 cells), and Hnf4α-Foxa1-Fos-Yap1 (n = 10,507 cells), profiled at day 20. Projection of fibroblast and iEP identity scores onto the UMAP embedding.
(E) Kernel density estimation of cell density for each reprogramming cocktail from (D).
(F) Violin plot of iEP identity scores for each reprogramming cocktail. ∗∗∗∗p < 0.0001, Wilcoxon test.
(G) Unsupervised cell type classification for each reprogramming cocktail, using normal and injured mouse liver as a reference. BEC, biliary epithelial cells. ∗p < 0.0001, randomized test.