Figure 2.
Transcriptomic enrichment analysis
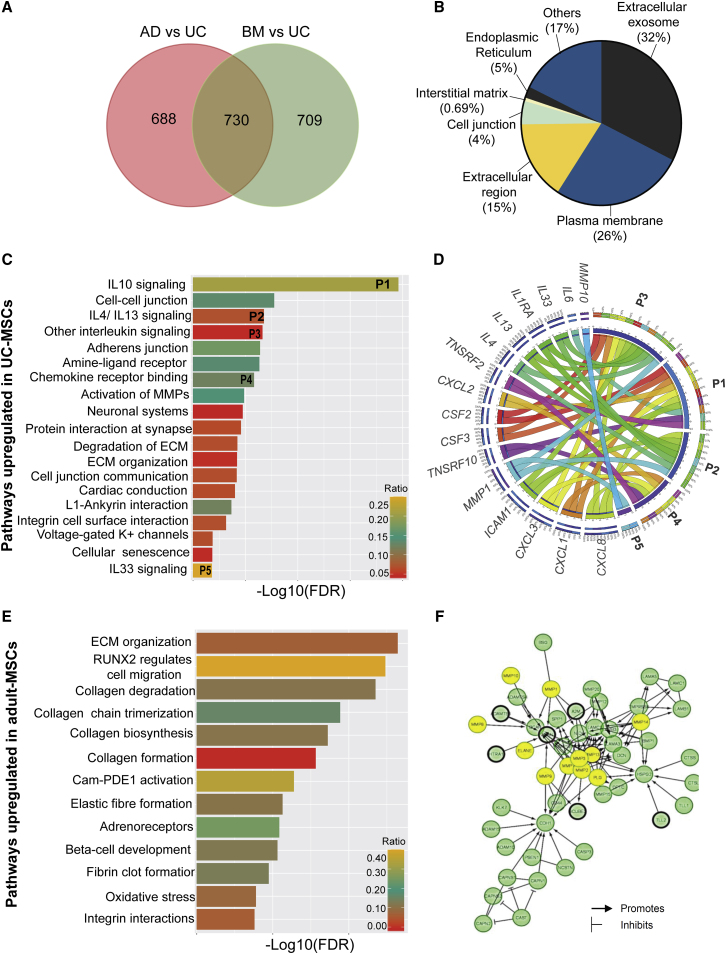
(A) Venn diagram showing the distribution of 2,127 DEGs in adult-MSCs compared with UC-MSCs.
(B) GO enrichment analysis indicates the cellular localization of the 730 common adult-MSC DEGs.
(C) All significant (FDR < 0.05) pathways upregulated in UC-MSCs compared with adult-MSCs.
(D) Circos plot depicts the relationship between enriched immune-regulatory pathways and their corresponding genes. Clockwise from top: pathways and genes are ordered by their number of interactions. Ribbon size encodes cell value associated with row/column segment pair. Column segment value and ribbon color are decided by the number of interactions.
(E) All significant (FDR < 0.05) pathways upregulated in adult-MSCs compared with UC-MSCs.
(F) Protein network of genes involved in extracellular matrix (ECM) organization pathway identified in adult-MSCs. Yellow circles indicate ECM proteolysis enzymes (MMPs, ELANE, and PLG) identified in our genomic screen. All other significant genes identified in our screen are highlighted with a black border. No fill circles indicate genes that are not validated experimentally.