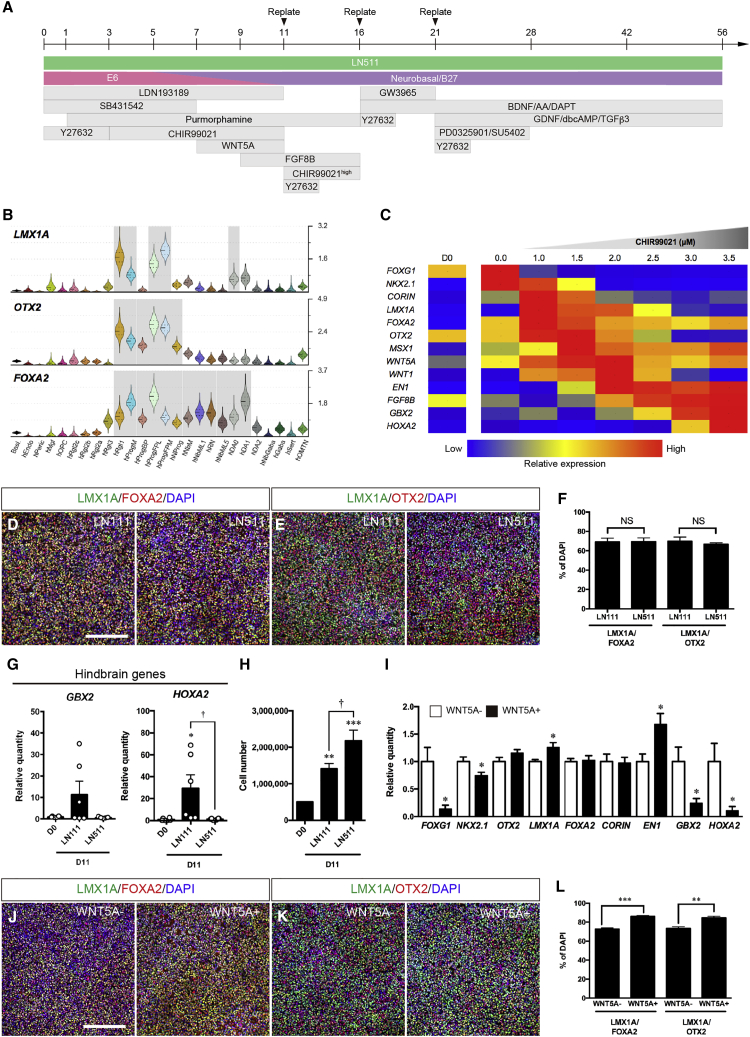
Figure 1.
Induction of floor plate progenitors from hESCs at day 11 using a new developmental-based protocol
(A) Schematic of the differentiation protocol for mDA neurons.
(B) Violin plots of LMX1A, FOXA2, and OTX2 generated from scRNA-seq data of developing human ventral midbrain shown across corresponding cell types. Right axis shows absolute molecular counts. Gray, enriched over baseline with posterior probability >99.8%. For cell type nomenclature, see La Manno et al. (2016).
(C) Gene expression profile of differentiated cells according to CHIR99021 concentration at day 11.
(D and E) Immunostaining of LMX1A+;FOXA2+ cells (D) and LMX1A+;OTX2+ cells (E) at day 11. Scale bar, 200 μm.
(F) Quantification of LMX1A+;FOXA2+ cells and LMX1A+;OTX2+ cells. NS, not significant (n = 4 independent experiments).
(G) qPCR analysis of GBX2 and HOXA2 in cells differentiating on either LN111 or LN511. ∗p < 0.05 versus D0; †p < 0.05 versus LN511 (n = 6 independent experiments).
(H) Cell yield by LN111 or LN511 at day 11. ∗∗p < 0.01, ∗∗∗p < 0.001 versus D0; †p < 0.05 versus LN511 (n = 6 independent experiments).
(I) qPCR analysis of differentiated cells in the presence/absence of WNT5A at day11 (n = 5–9 independent experiments). ∗p < 0.05 versus WNT5A(−) condition.
(J and K) Immunostaining of LMX1A+; FOXA2+ cells (J) and LMX1A+;OTX2+ cells (K) at day 11. Scale bar, 200 μm.
(L) Quantification of LMX1A+;FOXA2+ cells and LMX1A+;OTX2+ cells. ∗∗p < 0.01, ∗∗∗p < 0.001 versus WNT5A(−) condition (n = 4 independent experiments).