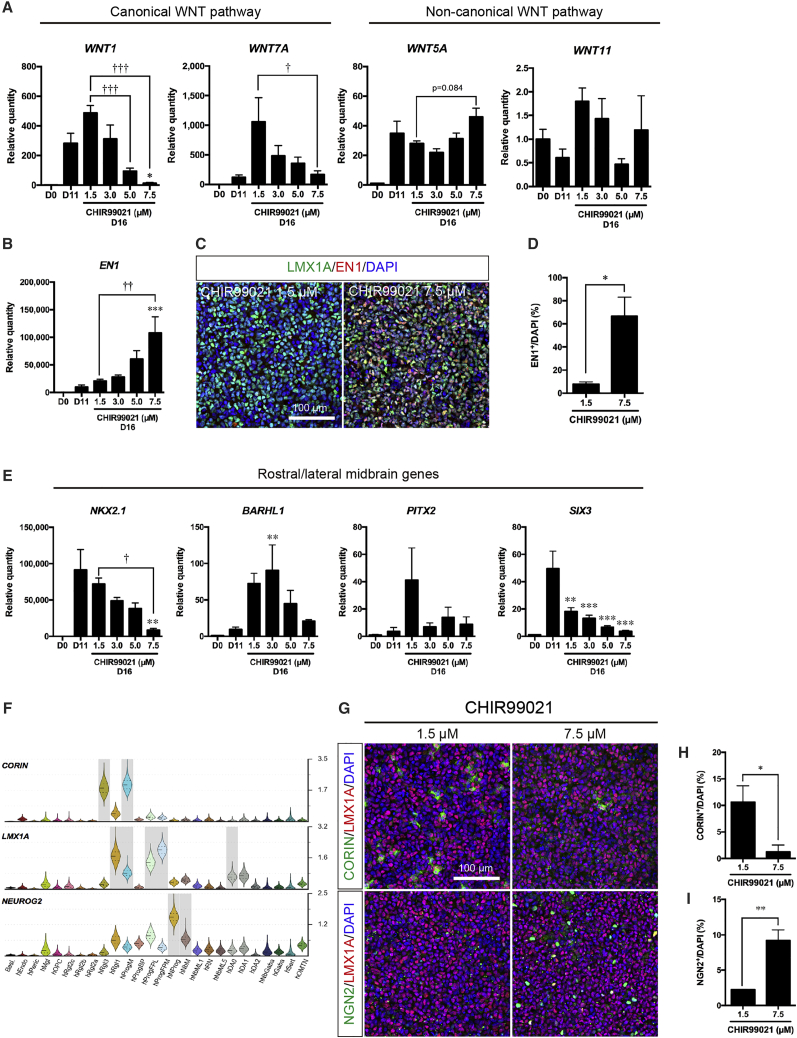
Figure 2.
Analysis of floor plate patterning in hESC-derived neural progenitors at day 16
(A) qPCR analysis of canonical WNT pathway and non-canonical WNT pathway at day 16. ∗p < 0.05 versus D11; †p < 0.01, †††p < 0.001 versus 1.5 μM CHIR99021 (n = 6 independent experiments).
(B) qPCR analysis of EN1 at day 16. ∗∗∗p < 0.001 versus D11; ††p < 0.01 versus 1.5 μM CHIR99021 (n = 6 independent experiments).
(C) Immunostaining of LMX1A+;EN1+ cells. Scale bar, 100 μm.
(D) Quantification of EN1+/DAPI+ cells. ∗p < 0.05 versus 1.5 μM CHIR99021 (n = 3 independent experiments).
(E) qPCR analysis of rostral and lateral midbrain markers at day 16. ∗∗p < 0.01, ∗∗∗p < 0.001 versus D11; †p < 0.05 versus 1.5 μM CHIR99021 (n = 6 independent experiments).
(F) Violin plots of CORIN, LMX1A, and NGN2 generated from scRNA-seq data of developing human ventral midbrain.
(G) Immunostaining of CORIN+;LMX1A+ cells and NGN2+;LMX1A+ cells. Scale bar, 100 μm.
(H and I) Quantification of CORIN+/DAPI+ cells (H) and NGN2+/DAPI+ cells (I). ∗p < 0.05, ∗∗p < 0.01 versus 1.5 μM CHIR99021 (n = 3 independent experiments).