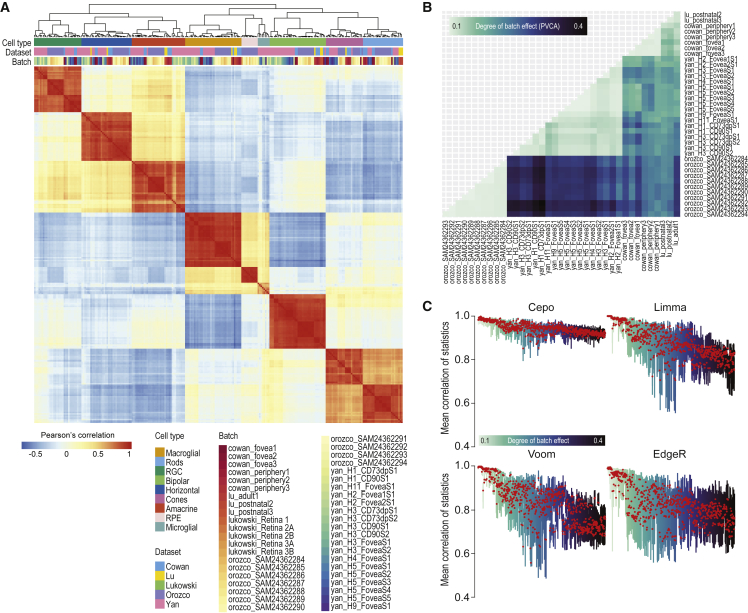
Figure 2.
Comparative analysis of cell-type-specific gene statistics across datasets and batches
(A) Correlation heatmap of cell-identity gene statistics generated from Cepo (Kim et al., 2021) for each cell type across datasets and batches. The heatmap is hierarchically clustered by the similarity of correlation profiles.
(B) Pairwise assessment of batch effect using principal variance component analysis (PVCA). The proportion of variance contributed by batch in each pair of datasets is visualized. A darker color denotes stronger batch effect.
(C) Boxplots of mean correlation of gene statistics from all retinal cell types for each pair of datasets illustrated in (B) using Cepo, Limma, Voom, and EdgeR statistics.