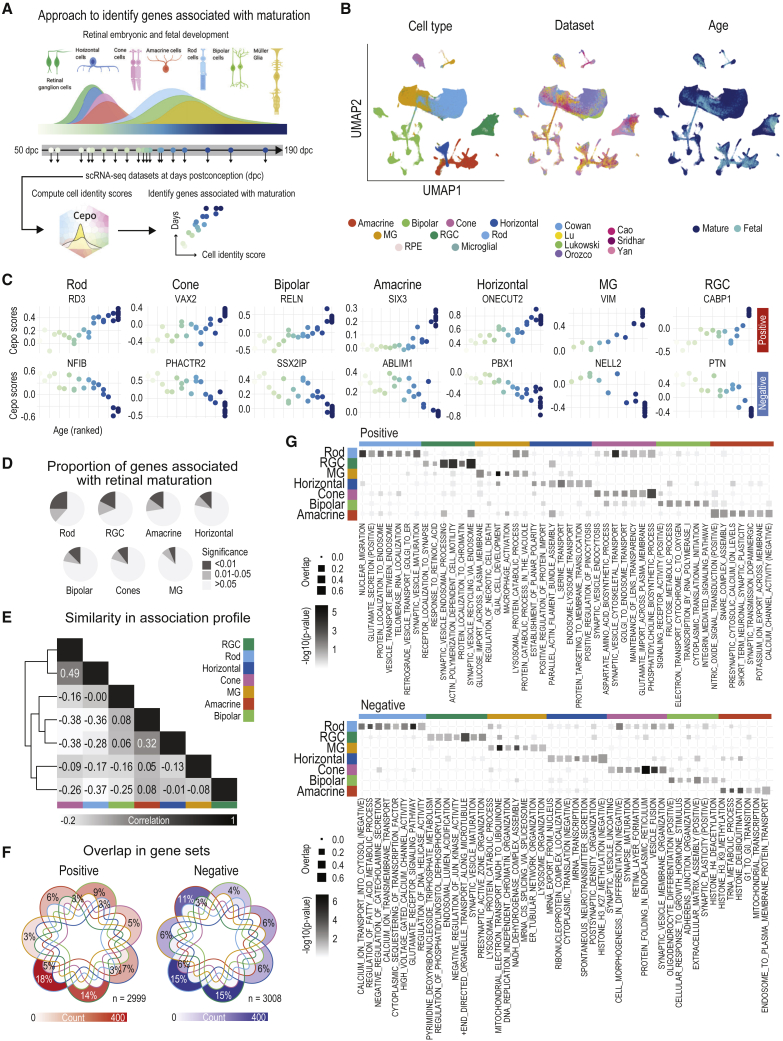
Figure 4.
The retinal cell-identity scores uncover genes associated with retinal maturation
(A) Schematic workflow to derive cell-type-specific maturation-associated genes. Scaling by day post-conception (dpc).
(B) UMAP of cell transcriptome profiles combining both adult and fetal tissues. Cells are colored by their type (left), dataset (middle), and sample type (right).
(C) Scatterplots of developmental age (x axis) and Cepo statistics (y axis). Points denote individual samples and are colored by their ranked developmental time point. The top and bottom show genes that are positively and negatively associated with age, respectively.
(D) Proportions of highly significant, significant, and insignificant maturation-associated genes for each cell type. Highly significant, false discovery rate [FDR]-adjusted p <0.01; significant, FDR-adjusted p between 0.01 and 0.05; and insignificant, FDR-adjusted p >0.05.
(E) Similarity in maturation association profiles between the cell types in terms of Pearson’s correlation coefficient.
(F) Overlap among the positively (left) and negatively (right) significant genes (FDR-adjusted p <0.05). The total percentage of overlap is highlighted for intersections greater than 2% of overlaps. The color scale denotes the absolute number of genes in each gene set.
(G) Enrichment of gene sets positively and negatively correlated with age.