Figure 6.
Benchmarking the human retinal organoid protocols in terms of their fidelity to the retinal tissue
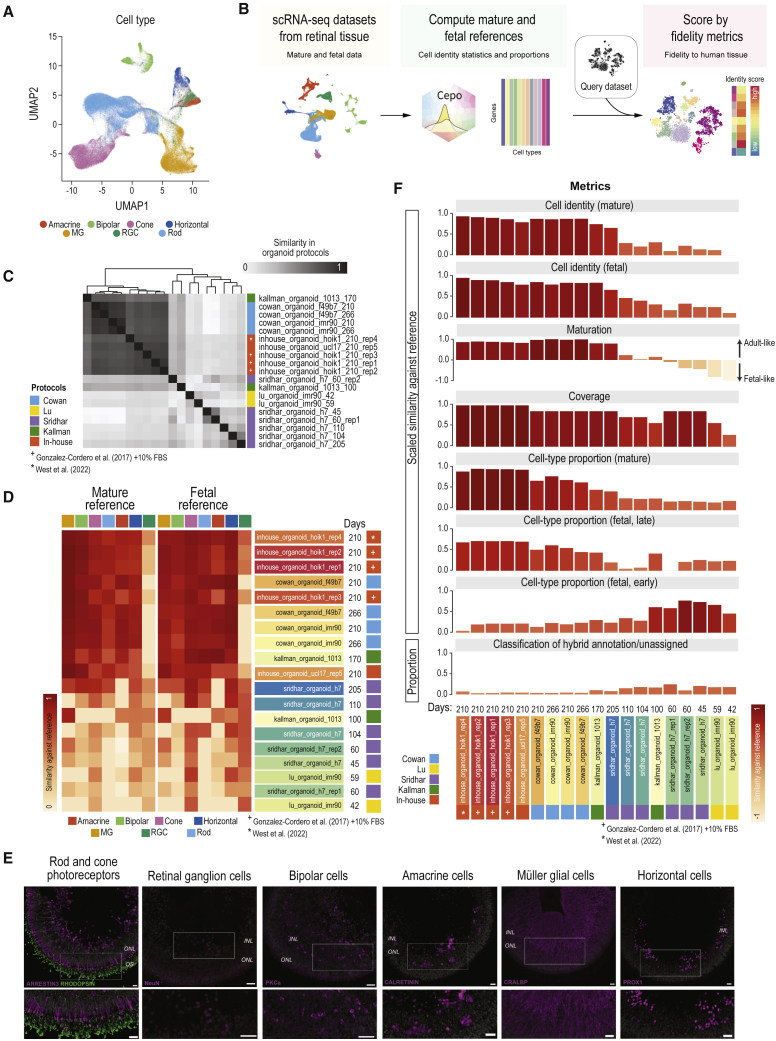
(A) UMAP of cell transcriptome profiles from human retinal organoids.
(B) Schematic of the benchmarking procedure and evaluation metrics.
(C) Similarity in cell-identity profiles between the scRNA-seq datasets generated from various organoid protocols. Similarity is measured in terms of Pearson’s correlation coefficient.
(D) Benchmarking results of the fidelity of the individual cell types to either the mature or the fetal reference. The cell-identity metric gives a scaled score between 0 and 1, where 1 denotes a high capacity and 1 denotes a low capacity of the protocol to generate cell types that closely match their tissue counterparts.
(E) Representative images of retinal cell types in human retinal organoids at 210 days of age. Abbreviations: ONL, outer nuclear layer; INL, inner nuclear layer; OS, outer segments. Scale bars: 20 μm.
(F) Final benchmarking results of the human retinal organoid protocols ranked by the combined score from six evaluation metrics: cell identity (mature), cell identity (fetal), maturation, coverage, cell-type proportion (mature), and proportion of potential off-target cells. The scores, except the maturation and proportion off-targets, have been scaled from 0 to 1 within each evaluation metric, where a value of 1 denotes a high capacity of the protocol to mimic the reference and 0 denotes low capacity.