Figure 4.
mRNA-seq analysis of HL-treated plants revealed a delayed HL response in tpt-2.
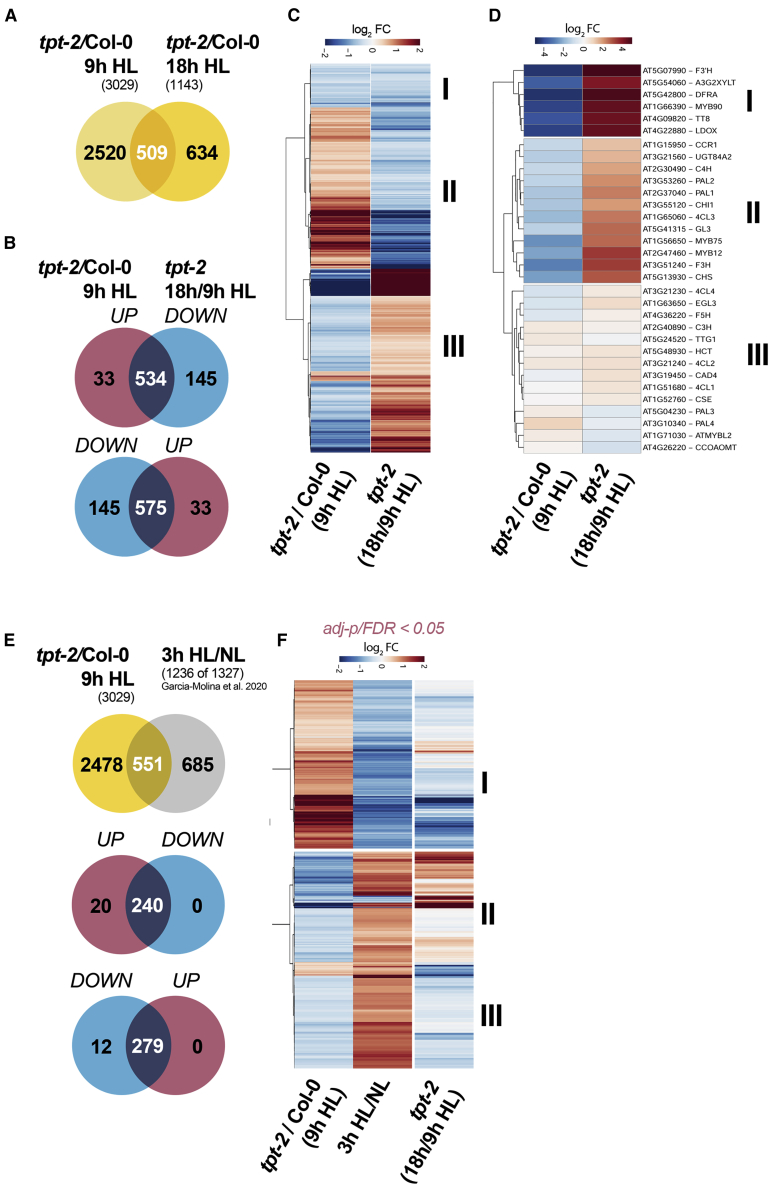
(A) Venn diagram depicting the overlap of significant DEGs (adjusted p < 0.05) between Col-0 and tpt-2 incubated for 9 h (red) and 18 h (blue) in HL.
(B and C) Venn diagram (B) and (C) heatmap showing transcripts differentially expressed in tpt-2/Col-0 at 9 h HL and their change in expression after 18 h relative to 9 h HL in tpt-2 (tpt-2 18 h/9 h HL) (adjusted p < 0.05). A significant number of up- or downregulated DEGs in tpt-2 relative to Col-0 after 9 h HL were induced or repressed in tpt-2 after 18 h of HL treatment (in total, 1109). DEGs in cluster I were downregulated in tpt-2/Col-0 after 9 h HL and were further repressed in tpt-2 18 h/9 h HL. Cluster II encompasses DEGs upregulated in tpt-2/Col-0 after 9 h HL but significantly downregulated in tpt-2 18 h/9 h HL. Cluster III contains DEGs downregulated at 9 h HL in tpt-2 relative to Col-0 but induced at 18 h HL treatment in tpt-2 relative to 9 h HL.
(D) Expression of genes encoding enzymes and transcriptional regulators of phenylpropanoid and flavonoid biosynthesis in tpt-2/Col-0 at 9 h HL and relative changes in tpt-2 at 18 h compared with 9 h HL. Clusters I and II contain transcripts that were strongly suppressed in tpt-2/Col-0 at 9 h HL but were induced after 18 h HL in tpt-2 relative to 9 h HL. Cluster III contains transcripts that showed only minor (relative) changes between 9 and 18 h HL treatment in tpt-2.
(E and F) Comparison of DEGs in tpt-2/Col-0 (9 h HL) with DEGs after a short-term (3 h) HL shift of Arabidopsis WT plants. DEGs after 3 h HL relative to NL conditions were extracted from Garcia-Molina et al. (2020) (FDR < 0.05). Of the 1327 DEGs after 3 h of HL treatment, transcripts for 1236 genes were found in the tpt-2 dataset and used for comparison (E).
(F) Heatmap comparing relative expression changes of transcripts in Col-0 treated with 3 h HL (Garcia-Molina et al., 2020), tpt-2/Col-0 (9 h HL), and tpt-2 (18 h HL/9 h HL). Changes in gene expression are given as log2 FC relative to the control, and hierarchical row clustering (Euclidean distance) using the ward.D method was used for all heatmaps.