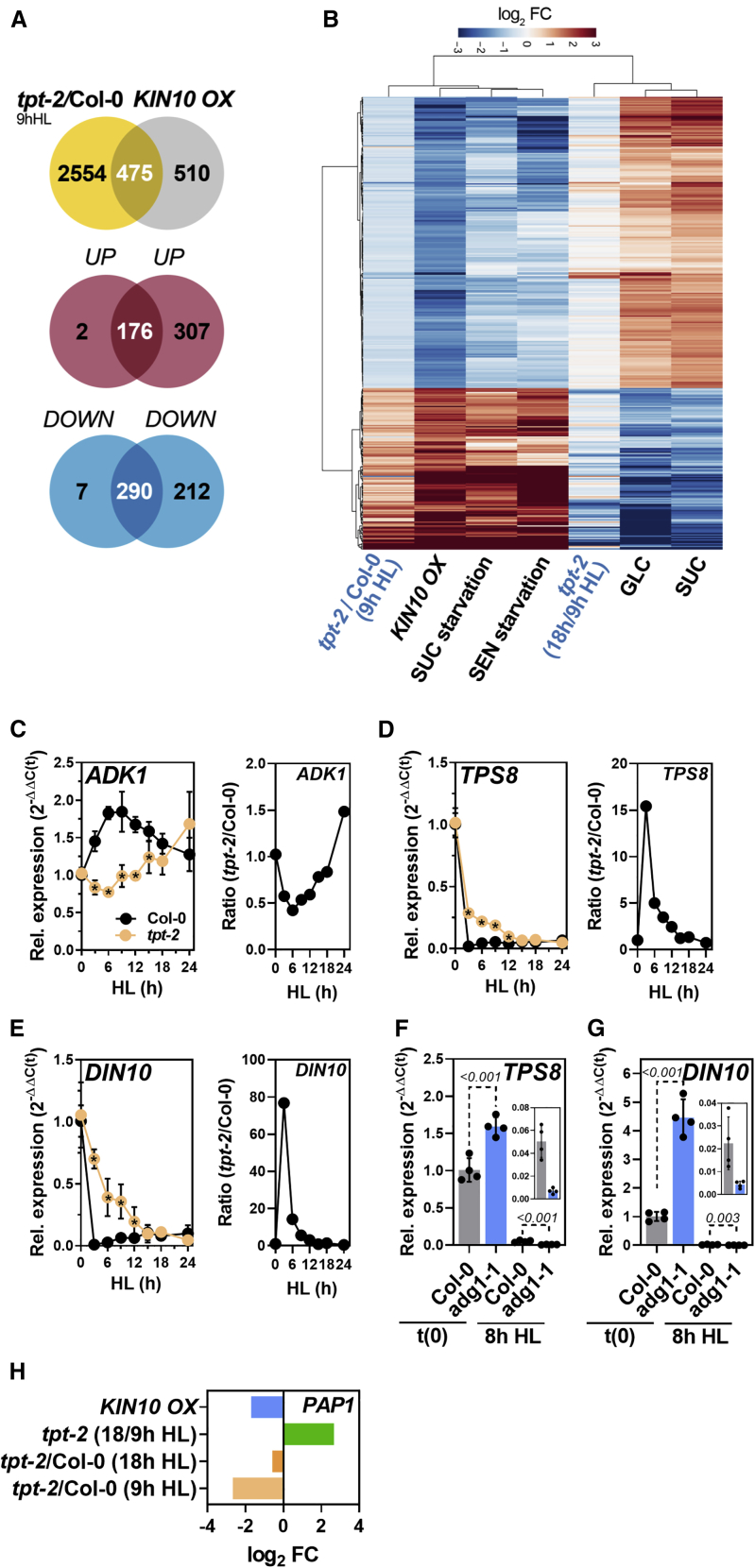
Figure 5.
Comparative transcriptome analysis indicated deregulation of SnRK1 signaling in tpt-2 during HL acclimation.
(A) Venn diagram showing the overlap between DEGs in tpt-2/Col-0 after 9 h HL and overexpression of KIN10 in protoplasts (KIN10 OX) extracted from Baena-Gonzalez et al. (2007) (Supplemental Data 7). Of the 985 reported transcripts for KIN10 OX, 475 were deregulated in the same direction in tpt-2 after 9 h HL (compare Venn diagrams for UP/UP and DOWN/DOWN).
(B) Heatmap comparing the relative expression of DEGs in tpt-2/Col-0 9 h HL (adjusted p < 0.05) and in tpt-2 (18 h/9 h HL) with the expression changes induced by KIN10 overexpression in protoplasts, SUC and senescence (SEN) starvation, GLC, and SUC feeding (Supplemental Data 7). Changes in gene expression are given as log2 FC relative to the control in each dataset. Hierarchical column and row clustering (Euclidean distance) using the ward.D method were applied.
(C–E) Relative expression of SnRK1-repressed (C)ADK1, and SnRK1-induced (D)TPS8 and (E)DIN10 during the HL shift kinetics of Col-0 and tpt-2.
(F and G) Expression of (F)TPS8 and (G)DIN10 in Col-0 and adg1-1. Gene expression was calculated using the 2−ΔΔCt method relative to Col-0 (t0, end of night) with SAND as the reference gene. Insets show the expression of TPS8 and DIN10 after 8 h HL. Statistical significance of differences between WT and tpt-2 at each time point was analyzed using Student’s t test (p < 0.05, asterisk inside the tpt-2 symbols) or 1-way ANOVA (Dunnett’s multiple comparisons test); adjusted p values are shown. Values are means ± standard deviations (n = 4).
(H) Expression of PAP1 in the indicated datasets/comparisons. Expression values for PAP1 in protoplasts overexpressing KIN10 were extracted from previously published data (Baena-Gonzalez et al., 2007). Changes in gene expression are given as log2 FC relative to the control in each dataset. PAP1 was significantly deregulated in KIN10 OX relative to control protoplasts, tpt-2/WT (9 h HL), and tpt-2 18 h/9 h HL.