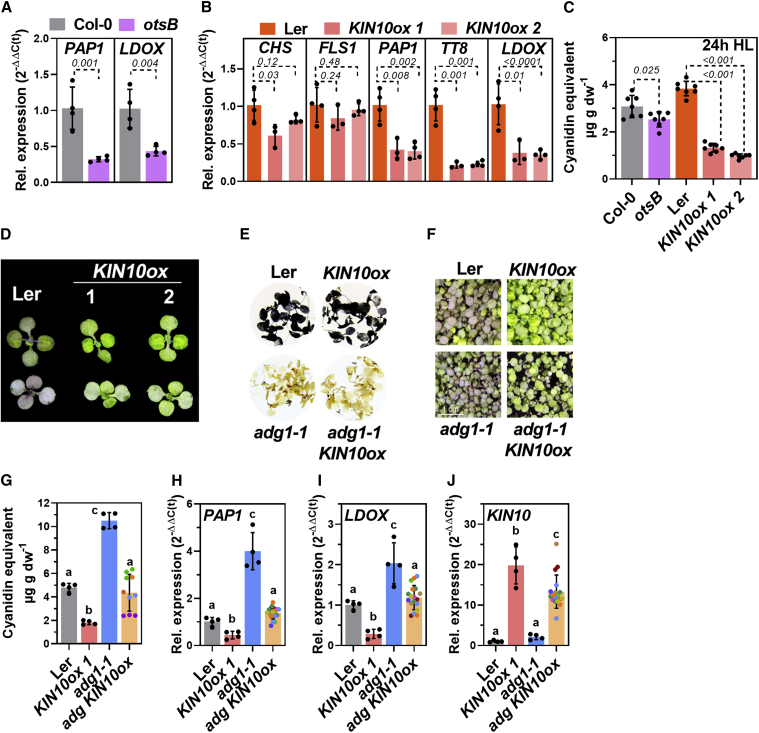
Figure 6.
Overexpression of KIN10 diminished HL-induction of FB
(A) Expression of PAP1 and LDOX in Col-0 and Col-0 expressing otsB (light purple) after 8 h HL.
(B) Expression of CHS, FLS1, PAP1, TT8, and LDOX in Landsburg erecta (Ler) (orange) and 2 independent overexpression lines for KIN10 in the Ler background (ox1 and ox2, pink and light pink) after 8 h HL. Plants were shifted at the end of the night. Gene expression was calculated relative to the corresponding WT background after 8 h HL using the 2−ΔΔCt method with SAND as the reference gene. Values are means ± standard deviations (n ≥ 3), and p values for Student’s t test between mutant and WT background are shown.
(C) Anthocyanin accumulation (expressed as cyanidin equivalents) in the lines shown in (A) and (B) after 24 h HL determined from 2 independent HL shift experiments. Values are means ± standard deviations (n = 7 from 2 independent shifts), and p values for Student’s t test between the mutant and the corresponding WT background are shown.
(D) Phenotypes of Ler, KIN10ox #1, and #2 after 24 h HL. Top: adaxial; bottom: abaxial.
(E and F) Staining of starch in seedlings exposed to 24 h HL (E) and (F) phenotypes of Ler, KIN10ox 1, adg1-1, and the adg1-1 KIN10ox double mutant after 24 h HL.
(G–J) Anthocyanin content after 24 h HL (G) and (H)PAP1, (I)LDOX, and (J)KIN10 expression after 8 h HL in the different genotypes. Note that for adg1-1 KIN10ox, seeds from different plants obtained after crossing and selection were used (same color of circle, see section “methods”). Each data point (circle) represents the result from 10–15 seedlings per “line” analyzed as 1 sample. Gene expression was calculated relative to Ler using the 2−ΔΔCt method with SAND as the reference gene. Values are means ± standard deviations. In (G)–(J), for Col-0, KIN10ox 1, and adg1-1, n = 4. For adg1-1 KIN10ox in (G), n = 10, and in (H)–(J), n = 18. Statistical significance of differences was analyzed using 1-way ANOVA (Tukey’s multiple comparisons test), and letters indicate significant differences at p < 0.05.