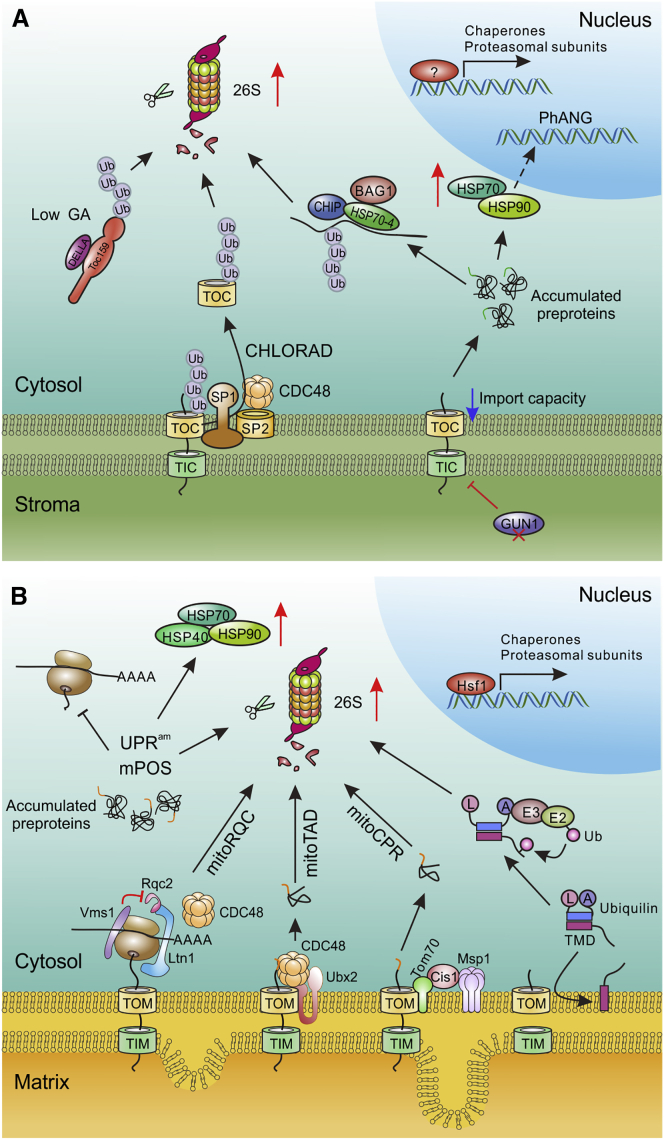
Figure 2.
Precursor accumulation response of chloroplasts and mitochondria.
(A) Chloroplast precursor accumulation response (PARcp). Impaired protein import into chloroplasts causes overaccumulation of chloroprotein precursors in the cytosol. In the Arabidopsis Toc159 mutant ppi2, the cytosolic HSP70 chaperone cyHSP70-4 (HSP70-4) and the E3 ubiquitin ligase CHIP were shown to participate in the degradation of overaccumulated chloroprotein precursors. The nucleotide exchange factor of HSP70, BAG1, recruits the proteasome to the substrates for degradation. Overaccumulation of chloroprotein precursors upregulates the cytosolic chaperones cyHSP70 and cyHSP90 and components of the 26S proteasome in gun1 mutants (which reduces plastid protein import capacity when chloroplast biogenesis is blocked, e.g., upon lincomycin or norflurazon treatment) to maintain cytosolic proteostasis. The chloroplast-associated protein degradation (CHLORAD) pathway is involved in the UPS-dependent turnover of components from the TOC complex during development and in response to stress. In the CHLORAD pathway, TOC subunits are ubiquitinated by the E3 ligase SP1 and extracted from the OEM by AAA+ ATPase CDC48 and Omp85 protein SP2 for proteasomal degradation. The DELLA protein RGL2 also participates in UPS-dependent TOC159 degradation at the early stage of germination when the GA concentration is low. However, whether the CHLORAD and DELLA pathways are involved in PARcp for degradation of clogged precursors in the import channel and/or overaccumulated precursors in the cytosol remains to be determined.
(B) Mitochondrial precursor accumulation response (PARmt). Multiple pathways have been identified in yeast, which surveils mitochondrial protein import and eliminates overaccumulated precursors in the cytosol. mitoRQC (ribosome quality control pathway for mitochondrial polypeptides): Listerin1 (Ltn1) gains access to precursors that stall in the ribosome, followed by ubiquitination, CDC48-mediated extraction, and proteasomal degradation. Vms1 (a 60S ribosome-binding protein) recognizes those stalled precursors that fail to be ubiquitinated by Ltn1 in close proximity to the translocase of outer membrane (TOM) complex at the OEM and prevents them from being CAT-tailed (C-terminal alanyl/threonyl sequences that mediate the aggregation of stalled polypeptides after import into mitochondria) by Rqc2. These precursors are imported into the mitochondrial matrix and either folded or degraded by the mitochondrial quality control system. mitoTAD (mitochondrial protein translocation-associated degradation): the mitoTAD pathway continuously monitors import under non-stress conditions to prevent clogging of the TOM channel by precursors. UBX domain protein Ubx2 (a component of ERAD) binds to the TOM complex to recruit CDC48 to extract the arrested precursors for proteasomal degradation. mitoCPR (mitochondrial compromised protein import response): inhibition of mitochondrial protein import induces the expression of Cis1, which associates with the TOM complex by interacting with Tom70. Cis1 recruits the AAA+ ATPase Msp1, which mediates the extraction and proteasomal degradation of arrested precursor proteins. Overaccumulation of mitochondrial precursor proteins (mPOS) in the cytosol causes an unfolded protein response activated by mistargeting of proteins (UPRam). mPOS and UPRam upregulate the expression of chaperones and proteasomal components through the action of heat-shock factor 1 (Hsf1) and inhibit translation to rebalance proteostasis. In mammals, ubiquilins bind to the transmembrane domain (TMD) of OEM-localized proteins in the cytosol to prevent their aggregation. The C-terminal ubiquitin-associating (UBA) domain (A) interacts with its N-terminal ubiquitin-like (UBL) domain (L) to stabilize the client–ubiquilin complex. Over time, the UBA domain recruits an unknown E3 ligase to ubiquitinate the substrates, while the free UBL domain directs the proteasomal degradation of the client proteins.