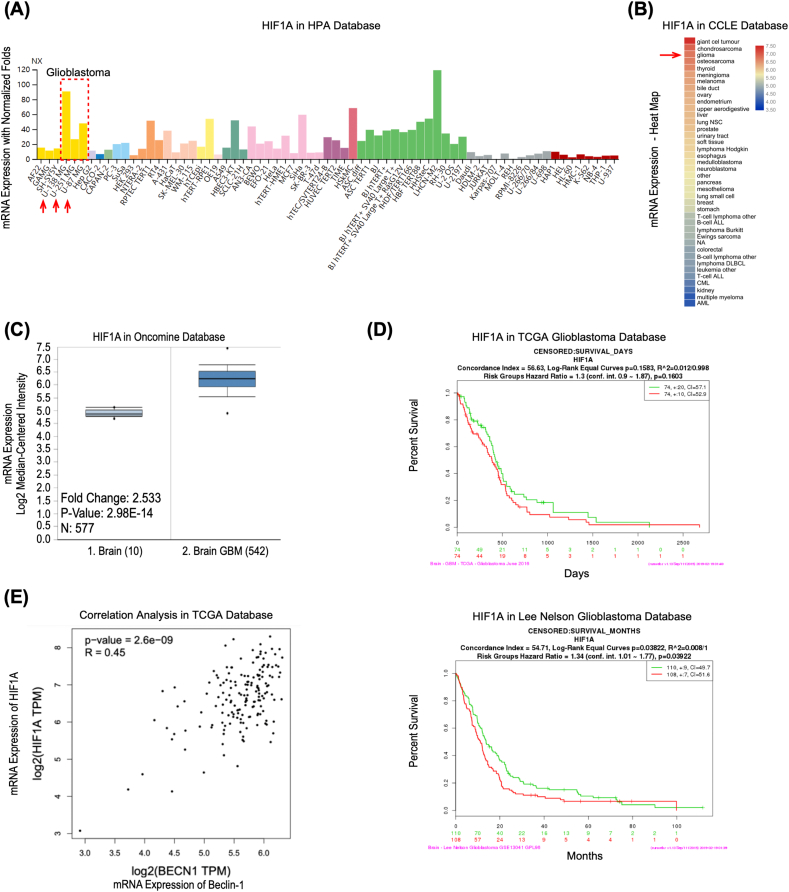
Figure 1.
HIF1A is overexpressed in GBM and predicts an adverse prognosis. (A) The mRNA expression level of HIF1A in different cell lines based on normalized RNA sequencing data on the Human Protein Atlas (HPA) database. https://www.proteinatlas.org. NX: Normalized expression, the expression level of gene–specific transcripts is given as normalized expression (NX) values, and transcripts with NX values ≥1 are considered as detected. The dotted box and red arrows indicate the Glioblastoma cell lines. (B) Heat map of the HIF1A mRNA expression level in various tissues based on the data in Cancer Cell Line Encyclopedia (CCLE). https://www.broadinstitute.org/ccle/home. GraphPad Prism plotted the heat map. The red arrow indicates the normalized mean value of mRNA expression in glioma tissue datasets. (C) HIF1A mRNA expression was significantly increased in brain glioblastoma tissue compared with normal brain tissue, as revealed by Oncomine data mining analysis (in the reporter 200,989 at probe set in the TCGA brain dataset). https://www.oncomine.org. (D) Multivariate Cox regression analysis of HIF1A gene expression and Kaplan–Meier survival curves for overall survival outcomes in the TCGA glioblastoma and Lee Nelson study datasets (The red and green lines correspondingly indicate a gene expression level above and below the median). https://cancergenome.nih.gov. (E) Correlation Analysis between HIF1A and Beclin–1 in the transcriptional level of clinical samples in TCGA GBM Tumor database, visualized by GEPIA website. https://gepia.cancer-pku.cn; TPM: transcripts per million, a normalized unit to measure the transcript coding gene expression.