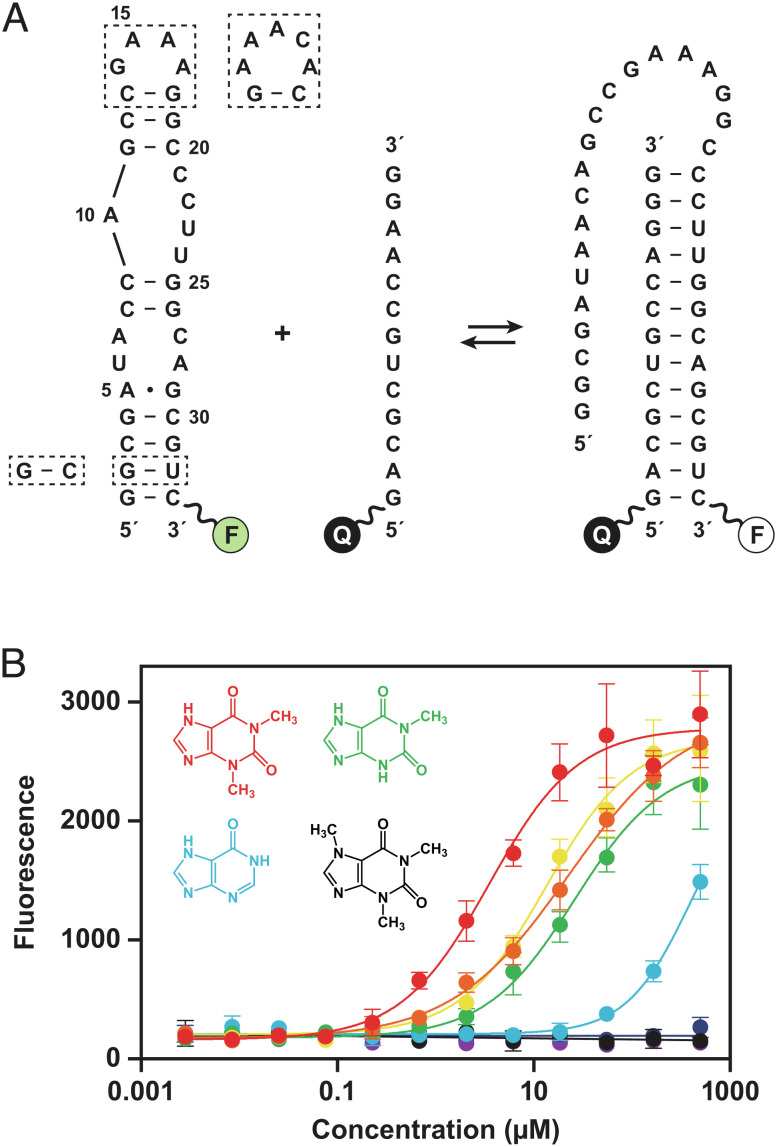
Fig. 1.
Strand invasion assay to screen for compounds that bind the theophylline aptamer. (A) Sequence and secondary structure of the theophylline aptamer, which was fluorescently labeled (F) at the 3′ end. Strand invasion by a partially complementary oligonucleotide, which has a fluorescence quencher (Q) at the 5′ end, results in decreased fluorescence. Two strategies were used to crystallize the aptamer, either replacing the GAAA tetraloop (nucleotides 14–17) with an AAACA pentaloop or replacing the G2-U32 wobble pair with a G–C Watson–Crick pair (dashed boxes). (B) Dose-dependent increase in fluorescence due to reduced propensity for strand invasion in the presence of theophylline (red), 1-carboxypropyl theophylline (orange), 3-methylxanthine (yellow), 1-methylxanthine (green), hypoxanthine (blue), 7-methylxanthine (indigo), 1,3-dimethyluric acid (violet), or caffeine (black). Values are based on four replicates, with error bars representing SE. The previously reported Kd values for binding of these compounds to the aptamer are 0.32, 0.93, 2.0, 9.0, 49, >500, >1,000, and 3,500 µM, respectively (18).