Significance
We found that the immunoregulatory receptor CTLA-4 is expressed on mTECs, where it mediates T cell central tolerance. This work deepens our understanding of how T cell tolerance is established and autoimmunity can arise and may inform the use of CTLA-4 fusion proteins and anti-CTLA-4 monoclonal antibodies employed clinically to treat various autoimmune diseases and cancers.
Keywords: Aire, CTLA-4, thymic epithelial cell, T cell, tolerance
Abstract
Medullary thymic epithelial cells (mTECs) are essential for the establishment of T cell central tolerance. The transcription factor Aire plays a key role in this process, but other factors remain understudied. We found that a small population of mTECs expressed the coinhibitory receptor cytotoxic T lymphocyte-associated protein 4 (CTLA-4). These CTLA-4+ cells were detectable in perinates, peaked around young adulthood and expanded sixfold in the absence of Aire. Single-cell transcriptomics revealed CTLA-4+ mTECs to express a distinct gene signature encoding molecules associated with antigen presentation and interferon-gamma signaling. Mice conditionally lacking CTLA-4 in thymic epithelial cells had no major immunological deficiencies but displayed a mildly increased inflammatory tone and a partial defect in the generation of Foxp3+CD4+ regulatory T cells. Consequently, these mice developed modest levels of autoantibodies and lymphocytic infiltration of peripheral tissues. Thus, CTLA-4 expression in mTECs complements Aire to establish T cell central tolerance.
Medullary thymic epithelial cells (mTECs) are essential for the establishment of T cell central tolerance because they ectopically express a diversity of peripheral-tissue antigens (PTAs), previewing the peripheral self to maturing T cells (reviewed in ref. 1). Through this process, self-reactive T cells can be deleted or diverted to the Foxp3+CD4+ regulatory T cell (Treg) lineage, averting autoimmunity. The importance of mTEC-driven central tolerance is illustrated by the phenotype of mice and humans with defects in mTEC differentiation and function, such as those caused by mutations in the nuclear factor kappa-B pathway and the transcription factor autoimmune regulator (Aire) (2, 3).
Aire, in particular, has been the focus of much attention because it plays a unique role in upregulating expression of numerous PTAs within mTECs, both directly in Aire-stage mTECs and indirectly in thymic mimetic cells (3, 4). While mice and humans with Aire-null mutations develop autoimmunity against Aire-induced antigens (3, 5, 6), the extent of autoimmunity varies widely across individual people and mouse strains (7–9). For example, most “Aire-deficient” mice on the nonobese diabetic background develop lethal autoimmunity, whereas Aire-deficient mice on the C57BL/6 (B6) background display only mild autoimmunity (9). Several explanations for this variation have been proposed, including genetic differences at Ctla4, Il2ra, and human leukocyte antigen loci, but they remain largely untested (7, 9)
Central T cell tolerance is complemented by a second tolerogenic mechanism, peripheral tolerance, which restrains autoreactive immunocytes that escaped into the periphery. Several mechanisms of peripheral tolerance have been described, some of the best characterized being cell-extrinsic inhibition by Tregs and cell-intrinsic inhibition by one of several cell-surface receptors. Cytotoxic T lymphocyte-associated protein 4 (CTLA-4; also known as CD152) is one such immunoregulatory receptor crucial for T cell tolerance. Mice lacking germline Ctla4 develop unrestrained lymphoproliferation and die within 6 wk of birth (10, 11). CTLA-4 binds costimulatory B7 molecules (CD80, CD86) on the surface of antigen-presenting cells (APCs) and functions through cell-intrinsic (reduction of costimulation) and cell-extrinsic (trogocytosis of B7 from APCs) mechanisms (12–16). However, the relative contributions of the various mechanisms to overall tolerance are unclear. Furthermore, although Tregs comprise one of the largest CTLA-4-expressing cell populations, constitutive conditional deletion of Ctla4 in Tregs only partially phenocopies germline Ctla4 deletion (17), and inducible conditional deletion in adult Tregs has almost no autoimmune phenotype whatsoever (18). Thus, while CTLA-4 is critical for restraining autoimmunity, the precise mechanisms by which it operates remain unresolved.
Here, we report that under normal conditions a small fraction of mTECs expressed CTLA-4. This subset was greatly increased in Aire-deficient mice. Conditional deletion of Ctla4 in mTECs revealed its role as a secondary regulator of T cell central tolerance.
Results
A Subset of mTECs Expresses CTLA-4 and Accumulates with Aire Deficiency.
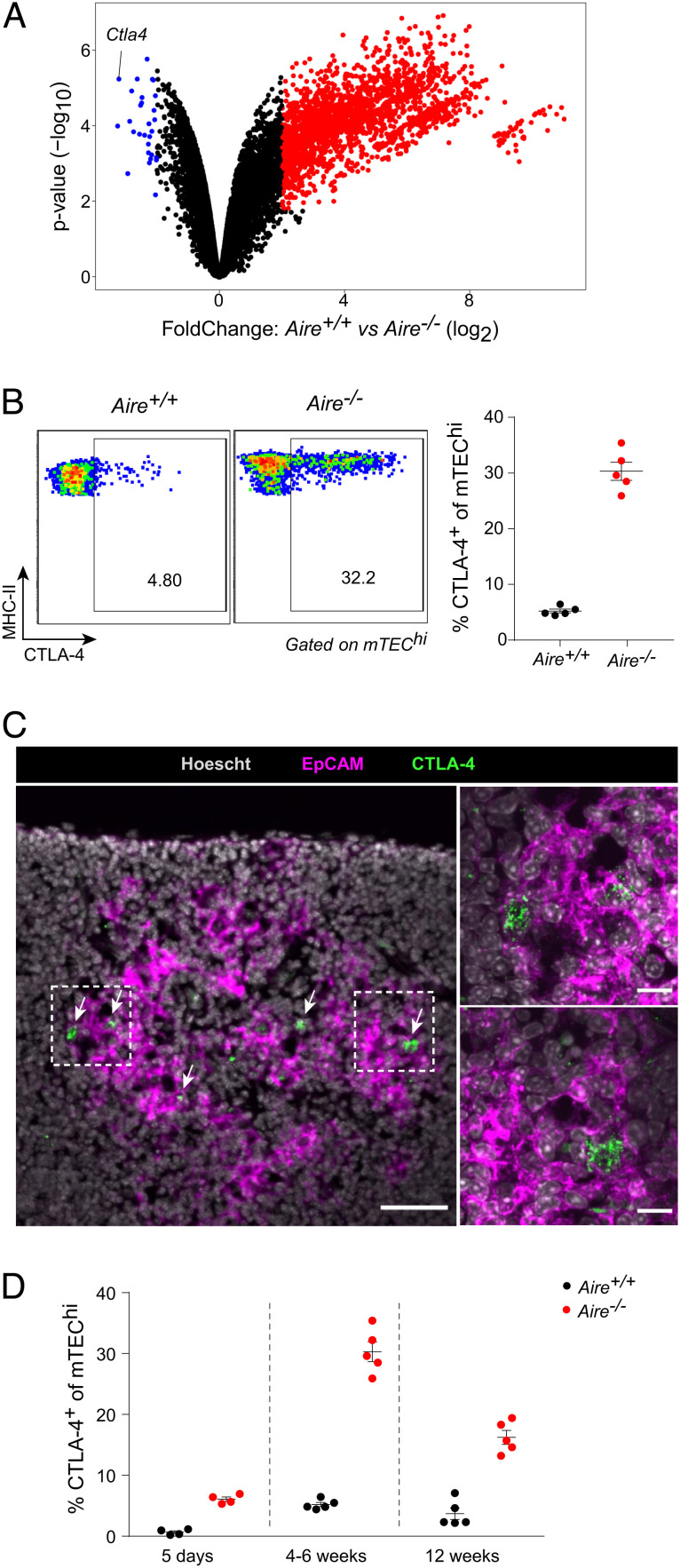
In analyzing published RNA sequencing (RNA-seq) data from mTEChi of B6.Aire+/+ vs. B6.Aire−/− mice (19), we observed that although many transcripts were downregulated in the absence of Aire, consistent with its role in PTA induction, only a few transcripts were substantially upregulated. One of the most highly induced of these transcripts was Ctla4 (Fig. 1A). Analysis of several similar RNA-seq datasets confirmed that this phenomenon was reproducible across experiments and laboratories (SI Appendix, Fig. S1A) (20, 21), and analysis of mTECs from Aire+/+ vs. Aire−/− mice on the BALB/c genetic background indicated that it was not unique to B6 mice (SI Appendix, Fig. S1B) (22). Another group recently reported CTLA-4 expression on mTECs as well (23).
Fig. 1.
CTLA-4 is expressed on mTECs and upregulated in Aire−/− mice. (A) Volcano plot of bulk RNA-seq of Aire-expressing mTEChi from Aire+/+ vs. Aire−/− mice. Each dot represents one gene, and differentially expressed genes (false discovery rate [FDR] < 0.05) are highlighted in red and blue. Data were reanalyzed from (19). (B) Flow plots (Left) and summarized data (Right) of CTLA-4 levels on mTEChi from Aire+/+ (n = 5) vs. Aire−/− (n = 5) mice. Each dot represents one mouse, and data were pooled from two independent experiments. Bars represent mean ± SEM. (C) Immunofluorescent microscopy of thymic sections from Aire−/− mice for the indicated markers at low (Left) and high (Right, corresponding to boxed regions) magnifications. Arrows indicate CTLA-4+ cells. Left scale bar, 50 μm; right scale bars, 10 μm. Data are representative of at least two independent experiments. (D) Summarized data of CTLA-4 levels on mTEChi from Aire+/+ vs. Aire−/− mice at 5 d (n = 4 vs. n = 4), 4–6 wk (n = 5 vs. n = 5), or 12 wk (n = 5 vs. n = 5) of age. Each dot represents one mouse, and data were pooled from six independent experiments. Data for 4–6 wk mice were reproduced from (B). Bars represent mean ± SEM.
By flow cytometry, we routinely observed expression of CTLA-4 on about 5% of mTECs in 4–6 wk-old Aire+/+ mice (Fig. 1B and SI Appendix, Fig. S1C). Expression of Ctla4 was largely restricted to major histocompatibility complex (MHC) class II-high Aire-stage mTECs (mTEChi), as compared to pre- and post-Aire MHC class II-low mTECs (mTEClo) (SI Appendix, Fig. S1D) (4). In Aire−/− mice, the fraction of CTLA-4-expressing mTECs rose to about 30% of all mTEChi, and the level of CTLA-4 protein on CTLA-4+ mTECs, as measured by geometric mean fluorescence intensity (gMFI), increased as well (Fig. 1B and SI Appendix, Fig. S1E). By immunofluorescence, we could identify CTLA-4+EpCAM+ cells in the medullary regions of the thymus (Fig. 1C). High-magnification imaging showed these cells to express CTLA-4 in a punctate, vesicular staining pattern, consistent with CTLA-4’s known cellular localization in endosomes (Fig. 1C) (24, 25).
Because the size and function of the thymic epithelium change dynamically with age (26, 27), we assessed the fraction of CTLA-4+ mTECs in perinatal (5 d old), young adult (4–6 wk old), and mature adult (12 wk old) Aire+/+ and Aire−/− mice (Fig. 1D). CTLA-4+ mTECs were rare but detectable in Aire+/+ perinates and were already fractionally increased in Aire−/− perinates. The fraction of CTLA-4+ mTECs peaked in young adults, along with the peak of thymic size, and then diminished somewhat in mature adults, with Aire−/− mice again showing expansion of CTLA-4+ mTECs relative to Aire+/+ mice.
Thus, we found that a small proportion of mTEChi expressed CTLA-4 in Aire+/+ mice. This population was expanded across multiple ages in Aire−/− mice.
Ctla4+ mTECs Express Antigen-Presentation and Interferon (IFN)-Response Programs.
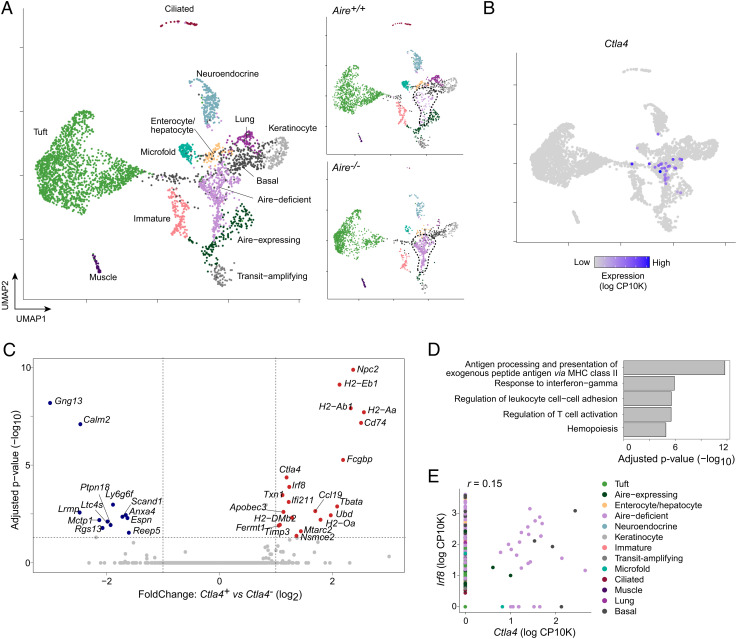
To better understand the nature of mTECs expressing CTLA-4, we analyzed published single-cell (sc) RNA-seq of mTECs from B6.Aire+/+ and B6.Aire−/− mice (4). Ctla4+ mTECs were readily detectable and grouped in a single cluster that was present at a low frequency in Aire+/+ mice and was greatly expanded in Aire−/− mice (Fig. 2 A and B), consistent with our cytofluorimetric characterization.
Fig. 2.
Ctla4+ mTECs display antigen-presentation and IFNγ signatures. (A) Uniform manifold approximation and projection (UMAP) plots of scRNA-seq of mTECs from Aire+/+ and Aire−/− mice, shown merged (Left) or split by genotype (Right). Each dot represents one cell, and dots are colored by cluster. The dashed lines indicate the “Aire-deficient” cluster. Data were reanalyzed from (4). (B) UMAP plot as in (A), colored by Ctla4 expression calculated as the natural log1p of counts per 10K total counts (log CP10K). (C) Volcano plot of gene expression in Ctla4+ vs. Ctla4− mTECs, as assayed by scRNA-seq. Each dot represents one gene, and differentially expressed genes (FDR < 0.05) are highlighted in red and blue. For (C–E), Ctla4+ mTECs were pooled from Aire+/+ and Aire−/− mice. (D) Pathway analysis displaying top gene ontology (GO) terms of differentially upregulated genes in Ctla4+ mTECs, ranked by significance. (E) Scatterplot displaying Irf8 and Ctla4 expression in individual cells, represented as dots. r, Pearson correlation.
Comparison of Ctla4+ and Ctla4− mTECs pooled from both genotypes revealed Ctla4 to be significantly associated with a distinct set of transcripts that included many related to antigen presentation by MHC-II molecules (H2-Aa, Cd74) and to the IFN response (Ifi211, Apobec3) (Fig. 2C). Ctla4 expression was also associated with expression of Irf8, encoding the transcription factor IRF8, which defines the chromatin program of Aire-deficient mTECs (4). Pathway analysis confirmed that Ctla4 expression was significantly associated with antigen presentation and with IFNγ-response programs (Fig. 2D); correlation analysis confirmed that the majority of Ctla4+ mTECs also expressed Irf8 (Fig. 2E). In contrast, Ctla4+ mTECs had lower levels of transcripts associated with terminal mTEC differentiation into mimetic cells, including Gng13, Calm2, and Ltc4s (Fig. 2C), suggesting that Ctla4+ mTECs were relatively depleted of terminal mTECs. Because Ctla4+ mTECs were more abundantly detected from Aire−/− mice (n = 28 cells), these analyses dominantly reflected the phenotype of Ctla4+ mTECs in the Aire−/− context; the low number of Ctla4+ mTECs detected in Aire+/+ mice (n = 4 cells) precluded direct analysis of transcriptional differences between Ctla4+ mTECs from Aire+/+ vs. Aire−/− mice.
In summary, Ctla4+ mTECs expressed a transcriptional program that was enriched in antigen presentation by MHC-II molecules and IFNγ-response pathways and that appeared to be controlled by IRF8. Ctla4+ mTECs showed a relative paucity of transcripts associated with terminally differentiated mTECs.
Mice Lacking CTLA-4 in mTECs Have a Perturbed Treg Compartment.
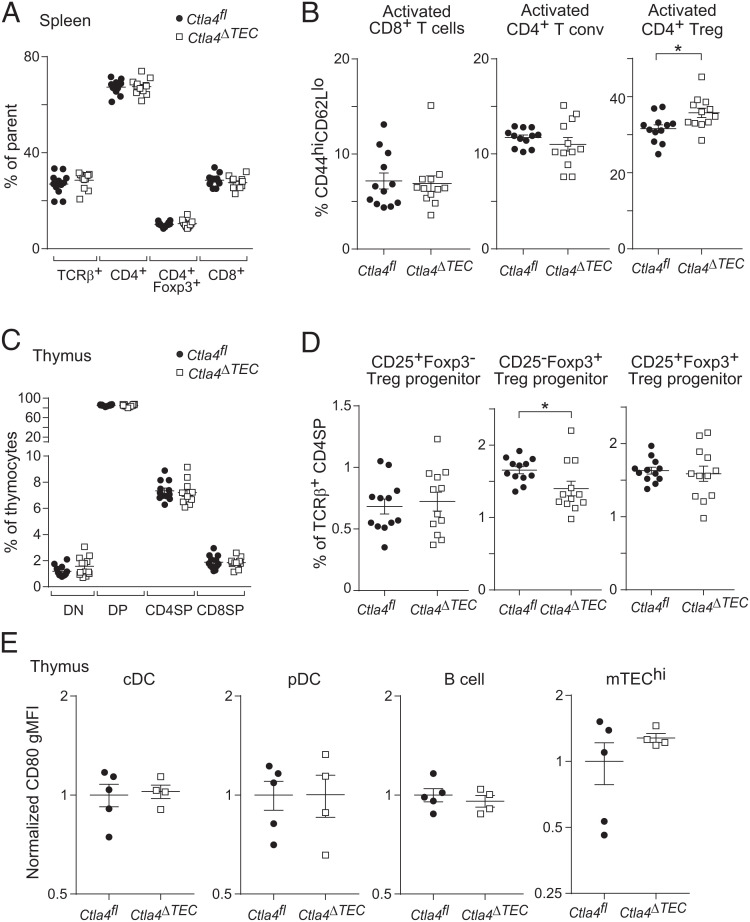
To understand the function of mTEC-expressed CTLA-4, we generated mice lacking CTLA-4 specifically in thymic epithelial cells by crossing in Foxn1cre and Ctla4flox genetic elements on the B6 background. Experimental mice were of the genotype Foxn1cre/+Ctla4flox/flox (Ctla4ΔTEC), while controls were littermates of the genotype Ctla4flox/flox (Ctla4fl). (Note that germline deletion of floxed alleles by Foxn1cre males rendered a breeding scheme with Cre-positive controls inviable (28)). Ctla4ΔTEC mice efficiently deleted CTLA-4 from mTECs (SI Appendix, Fig. S2A).
We first compared splenic T cells from Ctla4fl and Ctla4ΔTEC mice. There were no major perturbations in the number of total splenocytes, the fraction of T cells among splenocytes, or the distribution of T cells among the CD4+, CD8α+, and CD4+Foxp3+ compartments (Fig. 3A and SI Appendix, Fig. S2 B and C), nor were there significant increases in the activation status of conventional CD4+Foxp3− or CD8α+ T cells, as assessed by CD44 and CD62L expression (Fig. 3B). However, there was a modest but reproducible increase in the fraction of activated (CD44hiCD62Llo) Tregs (Fig. 3B), similar to what has been seen in other strains of mice with mild tolerance defects (29). This augmentation seemed to reflect an absolute increase in activated Tregs, as their number followed a similar trend, though this was not significant (SI Appendix, Fig. S3D). Thus, Ctla4ΔTEC mice did not manifest major T cell perturbations but showed signs of an elevated inflammatory tone that could be held in check by peripheral tolerance.
Fig. 3.
Mice lacking Ctla4 in mTECs have impaired thymic Treg generation and increased splenic Treg activation. (A–D) Summarized flow cytometry data for fractions of (A) splenic T cells, (B) activated (CD44hiCD62Llo) splenic T cells, (C) thymocytes, and (D) thymic Treg progenitors from Ctla4fl (n = 12) vs. Ctla4ΔTEC (n = 12) mice. In (A), TCRβ+ cells are gated on total splenocytes, while T cell subsets are gated on TCRβ+ cells. For all plots (A–D), each dot represents one mouse, data were pooled from two independent experiments, P-values were calculated by two-sided, unpaired Student’s t test, and bars represent mean ± SEM. (E) Summarized flow cytometry data for CD80 levels (geometric mean fluorescence intensity, gMFI) on thymic antigen-presenting cells from Ctla4fl (n = 5) vs. Ctla4ΔTEC (n = 4) mice. For each cell population, gMFIs are normalized to the mean of Ctla4fl controls. Each dot represents one mouse, data are representative of at least two independent experiments, and bars represent mean ± SEM.
We next compared thymic T cell subsets from mice of the two genotypes. Again, there were no significant changes in the distribution of major T cell subsets—namely, CD4/CD8 double-negative (DN), double-positive, and single-positive (CD4SP or CD8SP) thymocytes—or in the number of total thymocytes, suggesting that CTLA-4 ablation in mTECs did not grossly perturb thymic T cell maturation (Fig. 3C and SI Appendix, Fig. S3 E and F). However, there was a modest decrease in the fractional representation of the Treg progenitor expressing Foxp3 but not CD25 (Fig. 3D). This seemed to reflect an absolute decrease in CD25−Foxp3+ Treg progenitors, as their number followed a similar trend, though this was not significant (SI Appendix, Fig. S2G). The observed Treg progenitor perturbations suggested that the modest increase in inflammatory tone in Ctla4ΔTEC mice was secondary to a partial defect in Treg maturation.
To understand the mechanism by which CTLA-4 might affect Treg maturation, we analyzed levels of one of its ligands, CD80, on various thymic APC subsets, including conventional dendritic cells (DCs), plasmacytoid DCs, B cells, and mTECs. However, there were no significant changes in the levels of CD80 expressed by these various cell types (Fig. 3E). These results argue that mTEC-expressed CTLA-4 did not grossly diminish the amount of CD80 on thymic APCs but rather may have functionally inhibited costimulatory signaling.
Mice Lacking CTLA-4 in mTECs Develop Autoimmunity.
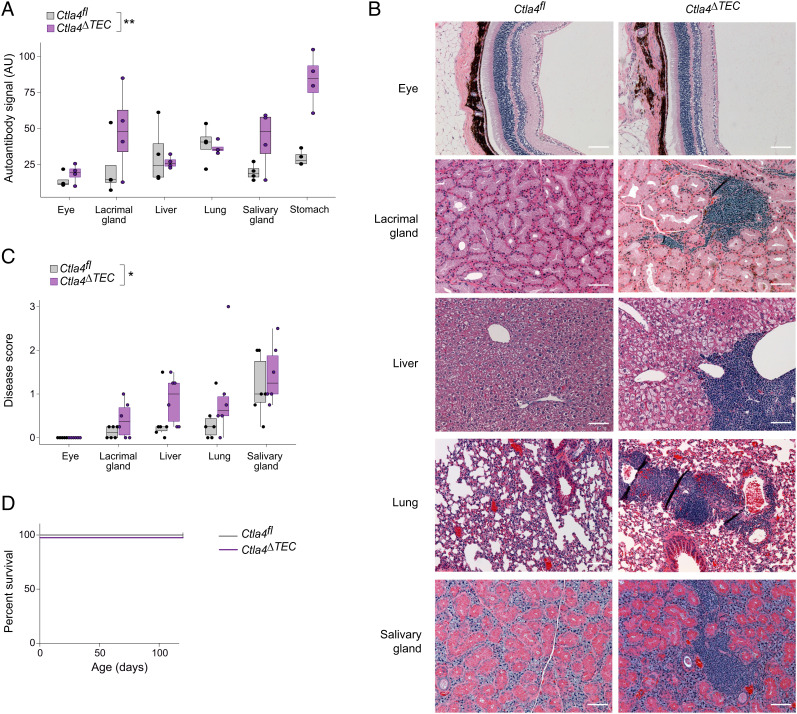
Finally, we asked whether mice lacking CTLA-4 in mTECs developed an autoimmune disease. We aged Ctla4fl and Ctla4ΔTEC mice for one year and collected serum for autoantibody analysis and peripheral organs for histological quantification of immunocyte infiltration and tissue destruction. Ctla4ΔTEC mice showed an increase in autoantibody burden across a variety of tissues, with enhanced oligoclonal autoantibody bands evident in the eye, lacrimal gland, salivary gland, and stomach, but not in the liver or lung. (Fig. 4A and SI Appendix, Fig. S3). The strength of these oligoclonal bands was comparable with that which we have observed in younger, 3–4-mo-old B6.Aire−/− mice (9). Ctla4ΔTEC mice also showed increased infiltration of several peripheral organs by immunocytes, albeit with a different tissue predilection, as the greatest infiltration was observed in the lacrimal gland, liver, and lung (Fig. 4 B and C). However, tissue architecture was generally preserved (Fig. 4B), and no animals suffered fatal autoimmunity during the study period (Fig. 4D), underscoring the mild nature of the tolerance defect in Ctla4ΔTEC mice. In the future, a systematic comparison of Ctla4ΔTEC mice to other autoimmune models will help contextualize the relative strength of autoimmunity observed here. Taken together with the cellular profiling, we conclude that thymic epithelial CTLA-4 played a minor but nonredundant role in the establishment of T cell central tolerance.
Fig. 4.
Mice lacking Ctla4 in mTECs develop mild autoimmunity. (A) Summarized quantification of autoantibodies in sera from 1-y-old Ctla4fl (n = 4) vs. Ctla4ΔTEC (n = 4) mice. Each dot represents one mouse; boxplots show median and interquartile range (IQR) as boxes and minimum and maximum values (up to ±1.5*IQR) as whiskers. The P-value for difference between genotypes was calculated by two-way ANOVA with genotype and organ as independent variables. (B) Representative hematoxylin and eosin (H&E) staining of tissue sections from the indicated organs from 1-y-old Ctla4fl (n = 6; n = 5 for eye) vs. Ctla4ΔTEC (n = 6) mice. (Scale bars, 100 μm.) (C) Summarized quantification of immunocyte infiltration and tissue destruction in tissue sections from (B). Each dot represents one mouse, and the boxplots and P-value were calculated as in (A). (D) Survival curves for Ctla4fl (n = 6) vs. Ctla4ΔTEC (n = 6) mice.
Discussion
In this study, we found that a small fraction of mTECs expressed CTLA-4 in wild-type mice and that this fraction was greatly expanded in the absence of Aire. This expansion was associated with antigen-presentation and IFNγ-responsive gene programs and appeared to serve as a secondary mechanism of central tolerance, as mice lacking CTLA-4 expression on mTECs had mildly impaired Treg production and modest increases in inflammatory tone, autoantibodies, and tissue infiltration.
We were surprised to see strong and tightly regulated CTLA-4 expression on mTECs, outside of its widely accepted localization on T cells. Notably, though most work on CTLA-4 has focused on its roles in T cells, a few studies have observed it to be expressed elsewhere (30–32). These results suggest that CTLA-4 expression might be a more generic mechanism than previously thought for modulating costimulatory signaling through B7, regardless of the cell type expressing CTLA-4. Within this framework, it will be of interest to identify the generic signals that induce CTLA-4 expression both on T cells and elsewhere, whether they be cytokines like IFNγ, as suggested here, or other factors.
Morimoto and colleagues also recently reported CTLA-4 expression on mTECs and its upregulation in Aire-deficient mTECs, in agreement with our results (23). Importantly, they used recombinant CTLA-4 (rCTLA-4) treatments and Ctla4 germline knockout mice to probe the mechanisms at play, while we generated mice with a conditional deletion of Ctla4 in thymic epithelial cells. Integration of these data yields a fuller picture of CTLA-4+ mTEC biology, especially when considering several points of difference between the studies. First, Morimoto et al. found that mice treated with presumably agonistic rCTLA-4 had diminished CD25−Foxp3+ Treg precursors, while we found that mice lacking CTLA-4 on mTECs also had diminished CD25-Foxp3+ Treg precursors. This somewhat paradoxical result suggests that CD28 signaling in maturing Tregs is tightly tuned by mTECs, and either too much or too little can have detrimental effects on Treg differentiation. Second, Morimoto et al. showed that mTEC-expressed CTLA-4 was capable of capturing B7 from thymic dendritic cells in vitro. But we did not observe a substantial diminishment of B7 on any thymic APC subset in Ctla4ΔTEC mice; this discrepancy might reflect a nontrogocytic functional role for CTLA-4 on mTECs in vivo and/or compensatory upregulation of B7 on APCs when CTLA-4 expression on mTECs is genetically ablated. Third, Morimoto et al. saw an alleviation of autoimmunity when Aire−/−Ctla4−/ thymi vs. Aire−/− control thymi were transplanted into nude mice, while we saw a mild exacerbation of autoimmunity when CTLA-4 was conditionally deleted in TECs on the Aire+/+ background, indicating that the status of other central tolerance mechanisms can dictate the protective vs. pathogenic role of CTLA-4 in mTECs.
Indeed, it is increasingly clear that multiple central and peripheral mechanisms integrate with Aire to enforce T cell tolerance (33–35). Aire plays a crucial role in ectopically inducing PTA expression; however, our scRNA-seq analysis demonstrated that loss of Aire was not neutral to mTECs but rather involved compensatory upregulation of a coordinated Aire-deficient gene program. We focused here on CTLA-4, but other factors, such as IL-13, were also upregulated in the absence of Aire and may play similar secondary roles in the induction of T cell tolerance. Additionally, recently described mimetic cell subtypes have unique expression profiles, including Il10 and Il25 in tuft mTECs and Ccl6, Ccl9, and Ccl20 in microfold mTECs, which may play functionally nonredundant roles in instructing thymocyte differentiation (36). Further studies will help clarify how the combination of these factors derived from heterogenous mTEC subsets integrate under homeostatic and pathologic conditions to ultimately control the T cell repertoire.
Materials and Methods
Gene-Expression Analyses.
Population-level and single-cell RNA-seq and microarray analyses were performed on published datasets using edgeR for bulk RNA-seq, Seurat for scRNA-seq, and R for microarray analyses. For single-cell analysis of Ctla4+ mTECs, pooled Ctla4+ mTECs from Aire+/+ and Aire−/− mice were used. Additional details are available in SI Appendix, Materials and Methods.
Mice.
Mouse work was performed at Harvard Medical School under specific-pathogen-free conditions using littermate controls. The strains used were C57BL/6J, B6.Aire−/−, B6.Foxn1-cre, B6.Ctla4-flox, and B6.Rag1−/−. Additional details are available in SI Appendix, Materials and Methods.
Isolation of mTECs. mTECs were isolated according to published protocols (4). Briefly, thymi were dissected, chopped, and incubated for 15 min in Dulbecco’s Modified Eagle Medium ( Gibco) plus 2% fetal calf serum ( Gibco), 25 mM 4-(2-hydroxyethyl)-1-piperazineethanesulfonic acid ( Lonza), 0.5 mg/mL collagenase (Sigma), and 0.1 mg/mL DNase (Sigma), then for 15 min in the same buffer plus 0.5 mg/mL collagenase/dispase (Roche) and 0.1 mg/mL DNase, and then briefly with 10 mM ethylenediaminetetraacetic acid. Cells were stained with primary antibodies against CD45, Ly51, MHC-II molecules, and/or CD80 (all BioLegend). In some cases, cells were fixed using fixation/permeabilization buffer (eBioscience) and stained with anti-CTLA-4 antibody (BioLegend) in permeabilization buffer (eBioscience).
Flow Cytometry.
Live cells were identified using 4′,6-diamidino-2-phenylindole or Fixable Yellow viability dye (Invitrogen). Cells were analyzed using LSRFortessa, LSR II, or FACSymphony instruments (all BD) at the Joslin Diabetes Center or Harvard Immunology Department Flow Cores.
Immunofluorescence Microscopy.
Thymi were dissected, fixed, frozen, sectioned, blocked, stained with conjugated primary antibodies against EpCAM and CTLA-4 (BioLegend), washed, counterstained with Hoescht 33342 (Sigma), mounted, and imaged by spinning disk confocal microscopy. Additional details are available in SI Appendix, Materials and Methods.
Isolation of Thymocytes and Splenocytes.
Thymi or spleens were dissected, passed over filters, and stained using primary antibodies against TCRβ, CD8α, CD25, CD44, CD45R, CD45, CD62L, CD80 (all BioLegend), CD4, and/or CD11c (both eBioscience). In some cases, cells were fixed using fixation/permeabilization buffer and stained with anti-Foxp3 antibody (eBioscience) in permeabilization buffer.
Autoimmune Monitoring, Serum Autoantibodies, and Histology.
Ctla4ΔTEC mice were observed weekly. Autoantibody and histology analyses were performed as previously described (3). Briefly, for autoantibody analysis, sera from individual mice were used at 1:500 dilutions for slot immunoblots of organ lysates from Rag1−/− mice. For histologic analysis, organs were fixed, embedded, sectioned, stained, and scored for immunocyte infiltration and tissue destruction by two blinded evaluators. Additional details are available in SI Appendix, Materials and Methods.
Statistics.
Sample sizes and statistical tests are noted in the figure legends. In general, flow cytometry data were compared by unpaired, two-sided Student’s t test in GraphPad Prism (v7.0), and histology and autoantibody data were compared by two-way ANOVA in R (v4.1.0). P < 0.05, *; <0.01, **; <0.001, ***; <0.00001, ****.
Supplementary Material
Appendix 01 (PDF)
Acknowledgments
We thank Dr. A. Sharpe for sharing the Ctla4flox mice; C. Araneo, D. Ischiu-Gutierrez, A. Wood, and A. Marotta at the Harvard Immunology Department and Joslin Diabetes Center flow cores; the Harvard Medical School Rodent Histopathology Core for histological tissue preparation; J. Lee, I. Magill, and A. Ortiz-Lopez for experimental help; K. Hattori for mouse help; L. Yang and B. Vijaykumar for computational help; C. Laplace for graphics; and T. Jayewickreme, M. Marin-Rodero, and K. Bansal for discussions. This work was supported by NIH grants AI088204 and DK060027 and a generous gift from the Howalt family (to D.M.) and NIH grant T32GM007753 (for D.A.M.).
Author contributions
D.A.M. and D.M. designed research; D.A.M. performed research; D.A.M. and D.M. analyzed data; C.B. and D.M. supervised research; and D.A.M., C.B., and D.M. wrote the paper.
Competing interests
The authors declare no competing interest.
Footnotes
Reviewers: L.J.B., University of Colorado - Anschutz Medical Campus; and J.V.R., The Rockefeller University.
Data, Materials, and Software Availability
Gene expression data were reanalyzed from published datasets: SRR2038194-SRR20381947, GSE180935, GSE53110, GSE194232 (bulk RNA-seq), GSE8564 (microarray), and GSE194252 (scRNA-seq). Scripts used for analyses are available at github.com/dmichelson.
Supporting Information
References
- 1.Abramson J., Anderson G., Thymic epithelial cells. Annu. Rev. Immunol. 35, 85–118 (2017). [DOI] [PubMed] [Google Scholar]
- 2.Shinkura R., et al. , Alymphoplasia is caused by a point mutation in the mouse gene encoding Nf-kappa b-inducing kinase. Nat. Genet. 22, 74–77 (1999). [DOI] [PubMed] [Google Scholar]
- 3.Anderson M. S., et al. , Projection of an immunological self shadow within the thymus by the aire protein. Science 298, 1395–1401 (2002). [DOI] [PubMed] [Google Scholar]
- 4.Michelson D. A., et al. , Thymic epithelial cells co-opt lineage-defining transcription factors to eliminate autoreactive T cells. Cell 185, 2542–2558 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Aaltonen J., et al. , An autoimmune disease, APECED, caused by mutations in a novel gene featuring two PHD-type zinc-finger domains. Nat. Genet. 17, 399–403 (1997). [DOI] [PubMed] [Google Scholar]
- 6.Nagamine K., et al. , Positional cloning of the APECED gene. Nat. Genet. 17, 393–398 (1997). [DOI] [PubMed] [Google Scholar]
- 7.Halonen M., et al. , AIRE mutations and human leukocyte antigen genotypes as determinants of the autoimmune polyendocrinopathy-candidiasis-ectodermal dystrophy phenotype. J. Clin. Endocrinol. Metab. 87, 2568–2574 (2002). [DOI] [PubMed] [Google Scholar]
- 8.Kuroda N., et al. , Development of autoimmunity against transcriptionally unrepressed target antigen in the thymus of Aire-deficient mice. J. Immunol. 174, 1862–1870 (2005). [DOI] [PubMed] [Google Scholar]
- 9.Jiang W., et al. , Modifier loci condition autoimmunity provoked by Aire deficiency. J. Exp. Med. 202, 805–815 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Waterhouse P., et al. , Lymphoproliferative disorders with early lethality in mice deficient in Ctla-4. Science 270, 985–989 (1995). [DOI] [PubMed] [Google Scholar]
- 11.Tivol E. A., et al. , Loss of CTLA-4 leads to massive lymphoproliferation and fatal multiorgan tissue destruction, revealing a critical negative regulatory role of CTLA-4. Immunity 3, 541–547 (1995). [DOI] [PubMed] [Google Scholar]
- 12.Linsley P. S., et al. , CTLA-4 is a second receptor for the B cell activation antigen B7. J. Exp. Med. 174, 561–569 (1991). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mandelbrot D. A., McAdam A. J., Sharpe A. H., B7–1 or B7–2 is required to produce the lymphoproliferative phenotype in mice lacking cytotoxic T lymphocyte-associated antigen 4 (CTLA-4). J. Exp. Med. 189, 435–440 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Krummel M. F., Allison J. P., CD28 and CTLA-4 have opposing effects on the response of T cells to stimulation. J. Exp. Med. 182, 459–465 (1995). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Qureshi O. S., et al. , Trans-endocytosis of CD80 and CD86: A molecular basis for the cell-extrinsic function of CTLA-4. Science 332, 600–603 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Schildberg F. A., Klein S. R., Freeman G. J., Sharpe A. H., Coinhibitory pathways in the B7-CD28 ligand-receptor family. Immunity 44, 955–972 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wing K., et al. , CTLA-4 control over Foxp3+ regulatory T cell function. Science 322, 271–275 (2008). [DOI] [PubMed] [Google Scholar]
- 18.Paterson A. M., et al. , Deletion of CTLA-4 on regulatory T cells during adulthood leads to resistance to autoimmunity. J. Exp. Med. 212, 1603–1621 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Meredith M., Zemmour D., Mathis D., Benoist C., Aire controls gene expression in the thymic epithelium with ordered stochasticity. Nat. Immunol. 16, 942–949 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sansom S. N., et al. , Population and single-cell genomics reveal the Aire dependency, relief from Polycomb silencing, and distribution of self-antigen expression in thymic epithelia. Genome. Res. 24, 1918–1931 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bansal K., et al. , Aire regulates chromatin looping by evicting CTCF from domain boundaries and favoring accumulation of cohesin on superenhancers. Proc. Natl. Acad. Sci. U.S.A. 118, e2110991118 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Venanzi E. S., Melamed R., Mathis D., Benoist C., The variable immunological self: Genetic variation and nongenetic noise in Aire-regulated transcription. Proc. Natl. Acad. Sci. U.S.A. 105, 15860–15865 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Morimoto J., et al. , Aire suppresses CTLA-4 expression from the thymic stroma to control autoimmunity. Cell Rep. 38, 110384 (2022). [DOI] [PubMed] [Google Scholar]
- 24.Linsley P. S., et al. , Intracellular trafficking of CTLA-4 and focal localization towards sites of TCR engagement. Immunity 4, 535–543 (1996). [DOI] [PubMed] [Google Scholar]
- 25.Alegre M. L., et al. , Regulation of surface and intracellular expression of CTLA4 on mouse T cells. J. Immunol. 157, 4762–4770 (1996). [PubMed] [Google Scholar]
- 26.Yang S., et al. , Regulatory T cells generated early in life play a distinct role in maintaining self-tolerance. Science 348, 589–594 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Baran-Gale J., et al. , Ageing compromises mouse thymus function and remodels epithelial cell differentiation. eLife 9, e56221 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Shi J., Getun I., Torres B., Petrie H. T., Foxn1[Cre] expression in the male germline. PLoS One 11, e0166967 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Dikiy S., et al. , A distal Foxp3 enhancer enables interleukin-2 dependent thymic Treg cell lineage commitment for robust immune tolerance. Immunity 54, 931–946 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Pistillo M. P., et al. , CTLA-4 is not restricted to the lymphoid cell lineage and can function as a target molecule for apoptosis induction of leukemic cells. Blood 101, 202–209 (2003). [DOI] [PubMed] [Google Scholar]
- 31.Stojanovic A., Fiegler N., Brunner-Weinzierl M., Cerwenka A., CTLA-4 is expressed by activated mouse NK cells and inhibits NK cell IFN-gamma production in response to mature dendritic cells. J. Immunol. 192, 4184–4191 (2014). [DOI] [PubMed] [Google Scholar]
- 32.Iwama S., et al. , Pituitary expression of CTLA-4 mediates hypophysitis secondary to administration of CTLA-4 blocking antibody. Sci. Transl. Med. 6, 230ra45 (2014). [DOI] [PubMed] [Google Scholar]
- 33.Proekt I., et al. , LYN- and AIRE-mediated tolerance checkpoint defects synergize to trigger organ-specific autoimmunity. J. Clin. Invest. 126, 3758–3771 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Benitez A. A., et al. , Absence of central tolerance in Aire-deficient mice synergizes with immune-checkpoint inhibition to enhance antitumor responses. Commun. Biol. 3, 355 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Policheni A. N., et al. , PD-1 cooperates with AIRE-mediated tolerance to prevent lethal autoimmune disease. Proc. Natl. Acad. Sci. U.S.A. 119, e2120149119 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Michelson D. A., Mathis D., Thymic mimetic cells: tolerogenic masqueraders. Trends Immunol. 43, 782–791 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Appendix 01 (PDF)
Data Availability Statement
Gene expression data were reanalyzed from published datasets: SRR2038194-SRR20381947, GSE180935, GSE53110, GSE194232 (bulk RNA-seq), GSE8564 (microarray), and GSE194252 (scRNA-seq). Scripts used for analyses are available at github.com/dmichelson.