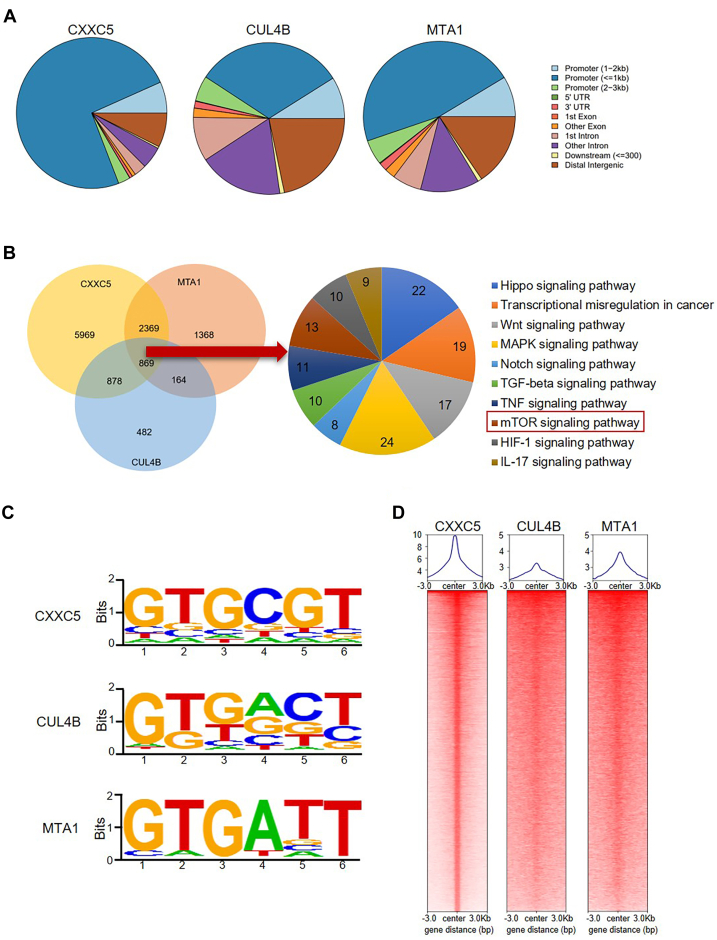
Figure 2.
Genome-wide identification of transcriptional targets for the CXXC5–CRL4B–NuRD complex.A, determination of the genomic distribution of CXXC5, CUL4B, and MTA1 binding sites by ChIP-Seq. B, a Venn diagram showing CXXC5, CUL4B, and MTA1 cobinding to promoters. After analysis, 869 overlapping target genes of CXXC5–CUL4B–MTA1 were obtained and clustered into functional groups. The detailed analysis results of ChIP-Seq have been summarized in Table S2. C, aanalysis of CXXC5-, CUL4B-, and MTA1-bound motifs using MEME suite. D, ChIP-Seq density heatmaps and profiles of CUL4B and MTA1 were analyzed against the CXXC5 binding sites as a background. ChIP-Seq, chromatin immunoprecipitation sequencing.