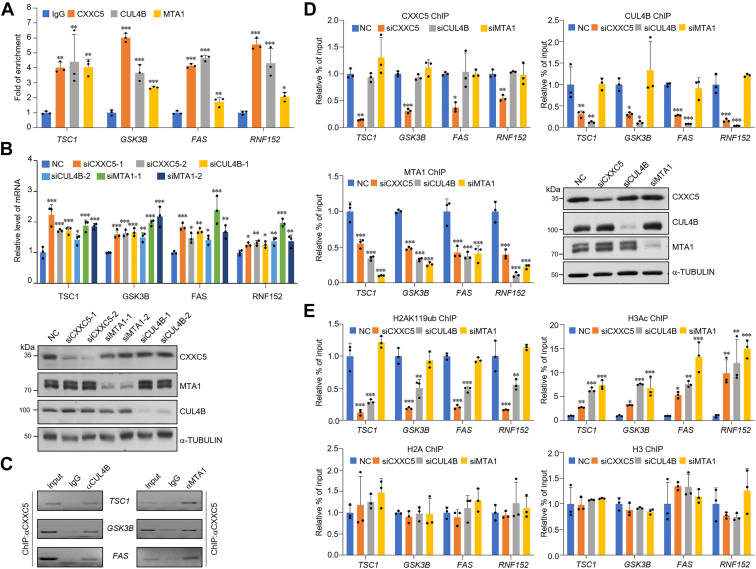
Figure 3.
Classification of downstream gene sets targeted by CXXC5-nucleated corepressor complexes.A, in MCF-7 cells, the ChIP-Seq results of the predicted genes with the indicated protein antibodies were verified by qChIP. The results show the calculation of fold change based on IgG. Each bar represents the mean ± SD from biological triplicate experiments. ∗p < 0.05, ∗∗p < 0.01, and ∗∗∗p < 0.001. B, MCF-7 cells were transfected with different groups of siRNAs to knockdown CXXC5, CUL4B, and MTA1, and the levels of representative downstream genes screened from ChIP-Seq were measured by RT–qPCR. Each bar represents the mean ± SD from biological triplicate experiments. ∗p < 0.05, ∗∗p < 0.01, and ∗∗∗p < 0.001. The knockdown efficiency of the proteins of interest was detected using Western blotting. C, ChIP/Re-ChIP experiments were performed for the promoters of the target genes with the proteins of interest antibodies. D, after transfection with the indicated siRNAs in MCF-7 cells, selected promoters were subjected to qChIP analysis in these cells using the indicated antibodies. Western blotting verified the knockdown efficiencies of CXXC5, CUL4B, and MTA1. Each bar represents the mean ± SD from biological triplicate experiments. ∗p < 0.05, ∗∗p < 0.01, and ∗∗∗p < 0.001. E, MCF-7 cells were transfected with siRNAs of the proteins of interest, and selected promoters were subjected to qChIP analysis in these cells using antibodies against histone modification, representing the function of the indicated proteins. Each bar represents the mean ± SD from biological triplicate experiments. ∗p < 0.05, ∗∗p < 0.01, and ∗∗∗p < 0.001. ChIP-Seq, chromatin immunoprecipitation sequencing; IgG, immunoglobulin G; qChIP, quantitative ChIP; qPCR, quantitative PCR.