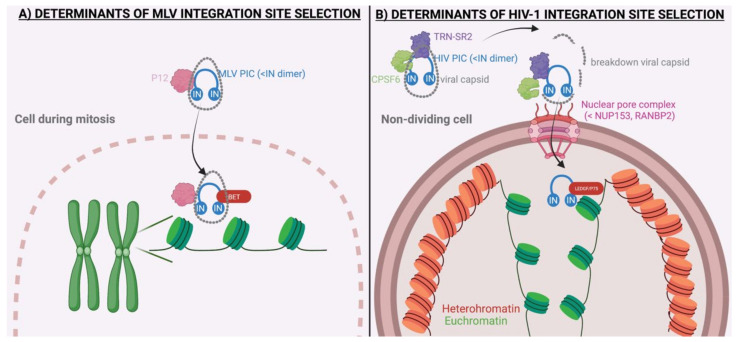
Figure 3.
(A) Determinants of the murine leukemia virus (MLV). MLV tends to integrate near transcription start sites (TSSs) and enhancers. The MLV p12 functions as a tether of the pre–integration complex (PIC) to the chromosomes, but its role in integration site selection is under debate. In contrast to HIV-1, MLV does not have an active nuclear entry route but is dependent on mitosis for nuclear entry. After nuclear entry and mitosis, bromodomain and extra–terminal motif (BET) proteins direct the integration pattern of MLV. BET proteins consist of an extra–terminal domain, which binds the MLV integrase (IN), and two bromodomains (BDI and BDII) that bin acetylated lysine in the nucleosomes. (B) Determinants of human immunodeficiency virus 1 (HIV-1). HIV-1 tends to integrate near decondensed chromatin, associated with active transcription, at the nuclear periphery. The HIV-1 pre–integration complex (PIC) can infect non–dividing cells by crossing the intact nuclear membrane via nuclear pore complexes. Nuclear import affects the HIV-1 integration pattern, with the ran–binding protein 2 (RANBP2, also called Nup358), transportin SR2 (TRN–SR2, also called transportin 3) and nucleoporin 153 (Nup153) as determinants of integration site selection. After nuclear import, lens epithelium–derived growth factor (LEDGF/p75) is considered the major contributing factor to integration site selection. LEDGF/p75 consists of an integrase–binding domain (IBD), binding the viral IN, and a PWWP domain, binding epigenetic marks in the genome associated with active transcription such as H3K36me2/3. In addition, the three–dimensional organization in the nucleus and the epigenetic landscape of chromatin affect HIV integration, as HIV-1 tends to integrate into less condensed euchromatin and avoids integration in closed heterochromatin regions. (Figure created with Biorender.com on 17 December 2022).