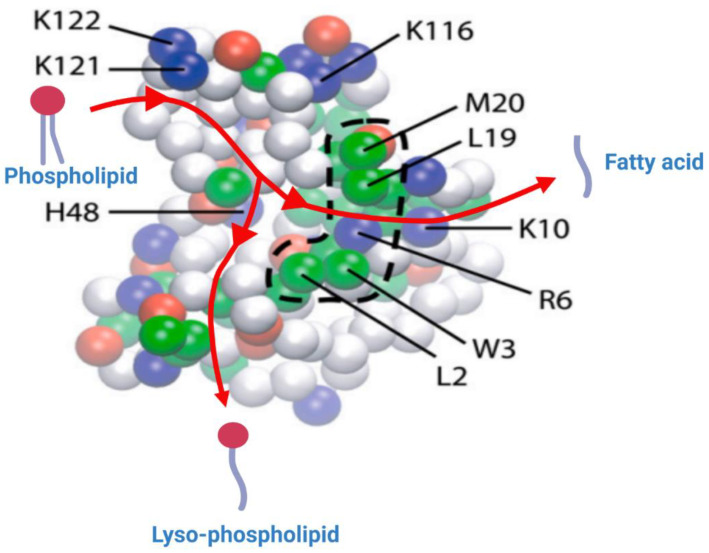
Figure 7.
Representation of the bottom (inter)face of the sPLA2, which remains in contact with the lipid bilayer, generated through the CG simulation. The amino acid residues which remained deep in the bilayer are represented in strong colors, and the ones that are away from the bilayer are shown in faded colors. The hydrophobic, basic, acidic and polar residues are shown as green, blue, red and white spheres, respectively. The broken line delimiting the cluster of hydrophobic residues (Trp-3, Leu-19, Met-20) highlights the key residues believed to anchor the sPLA2 into the lipid bilayer. The basic residues (Arg-6, Lys-10, Lys-116, Lys-121 and Lys-122) form electrostatic interactions with the phosphate group and with the carbonyl group of the lipid substrate. Reprinted/adapted with permission from [22]. Copyright 2008, Elsevier.