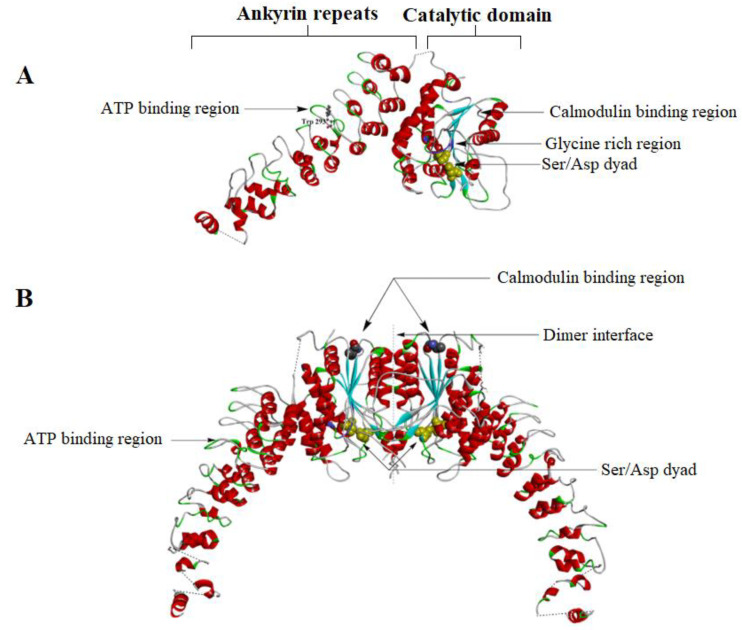
Figure 13.
(A) The crystal structure of the monomeric form of iPLA2β (PDB- 6AUN). The ankyrin repeats (ANK) are shown on the left side and the catalytic domain on the right side. The linker between the ANK and the CAT (connected by a dashed line) is unresolved due to less electron density. Near the 6th ankyrin repeats, there is the Trp293 residue which is believed to be the ATP binding site. The catalytic dyad (Ser465 and Asp598) is shown in purple in the catalytic domain. The glycine-rich region, which forms a rigid handle, is shown in blue. Earlier studies showed it as the ATP binding domain. The CaM binding region is near residue 630. (B) The dimeric structure of the iPLA2. One CaM molecule binds two iPLA2 molecules and stabilizes the close confirmation of the active sites of the enzyme. Note that the two active sites are located near the dimeric interface (Image generated using BIOVIA Discovery studio).