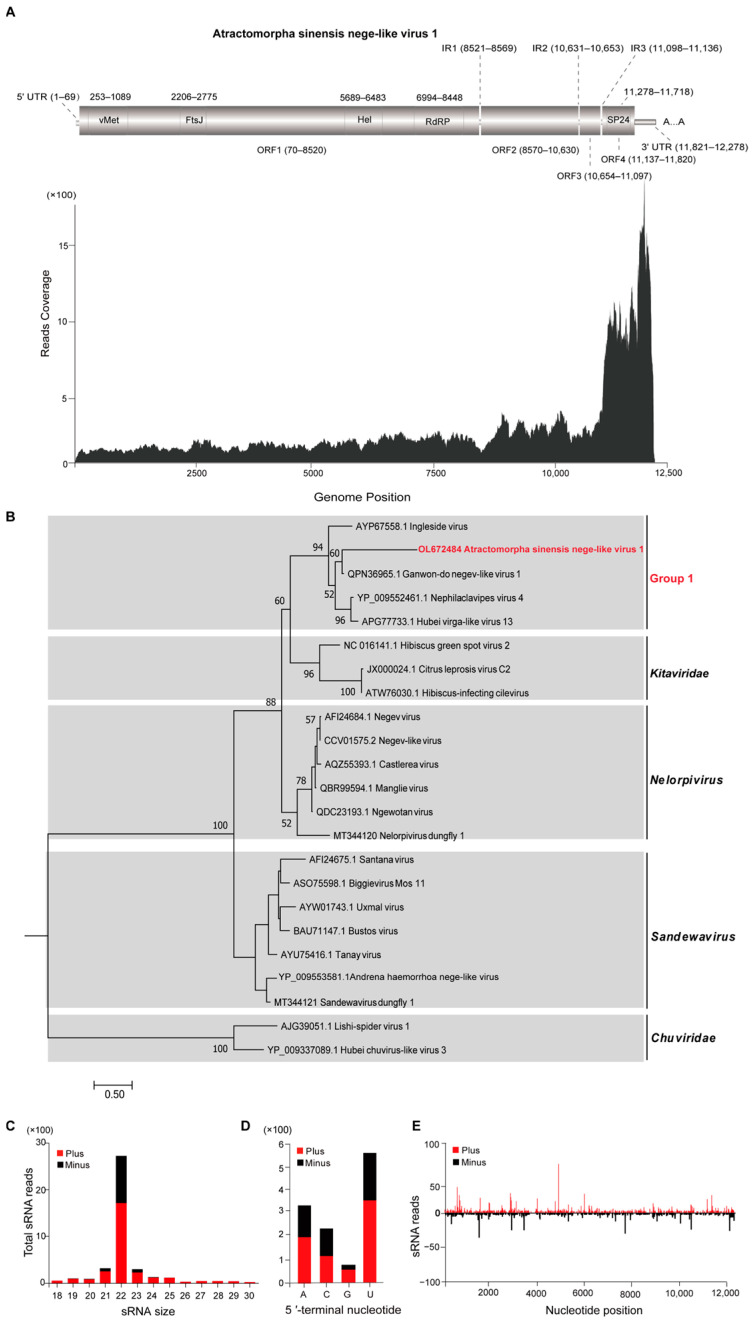
Figure 1.
Atractomorpha sinensis nege-like virus. (A) Genetic structure and transcriptome raw read coverage; vMet, viral methyltransferase domain; FtsJ, Ribosomal RNA methyltransferase; Hel, RNA virus helicase core domain; RdRP, RNA-dependent RNA polymerase domain; SP24, a putative virion membrane protein. (B) Phylogenetic analysis of ASNV1 and other negeviruses based on the conserved RdRP amino acid sequences. Bootstrap values are displayed over each node of the tree (when > 50), and scale bars represent percentage divergence, while the ASNV1 identified in this study is indicated with red fonts. (C) Size distribution of ASNV1 sRNAs. (D) Preference of 5′-terminal nucleotide. (E) Distribution of ASNLV1-derived sRNA along the corresponding viral genome.