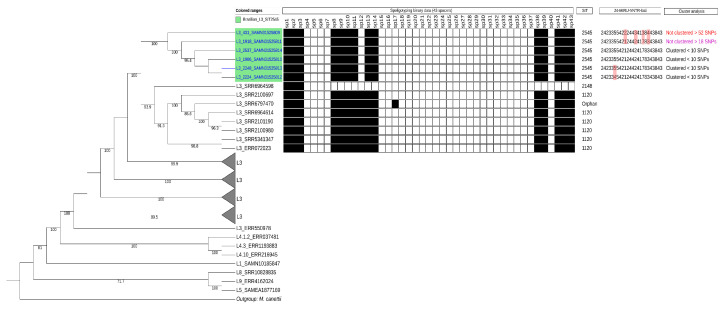
Figure 3.
Cladogram based on the phylogenetic reconstruction of single-nucleotide variants (SNV) determined by whole-genome sequencing analysis with the MTBseq pipeline. The four collapsed clades represent 1431 of the including 1350 Mycobacterium tuberculosis L3 genomes acquired from publicly available databases, an outgroup Mycobacterium canettii and other M. tuberculosis representative lineages human-adapted (L1–L9). The six L3 genomes from Brazil (green) at the nodes of the cladogram is annotated with their spoligotyping pattern, MIRU-VNTR and 12 SNV pairwise genetic distance comparisons. The L3 genomes of the closely related subclade has a different spoligotyping pattern than Shared International Type (SIT) 2545.