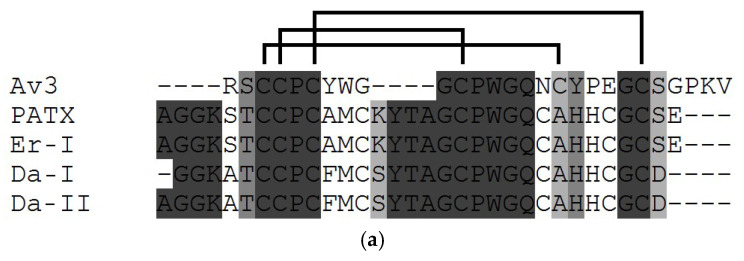
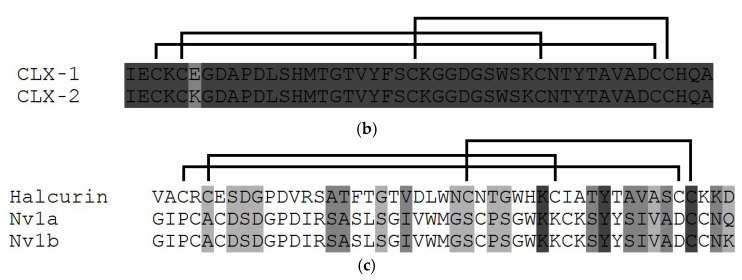
Figure 4.
Multiple alignment of the amino acid sequences of: (a) type 3 sea anemone NaTxs: ATX-III (=Av3) (UniProt ID: P01535) from A. sulcata [124], PaTX (P09949) from E. quadricolor (=P. actinostoloides) [126], Er-I (P09949/P09949-1) from E. ramsayi, Da-I (a major form) and Da-II (P0DMZ2/P0DMZ2-1) from D. armata [127]; (b) type 4 NaTxs CLX-1 (UniProt ID: P14531) and CLX-2 (P49127) from C. parasitica [132]; and (c) Halcurin (UniProtKB: P0C5G6) from H. carlgreni [135] and Nv1 (B1NWS1) from N. vectensis [137]. The sequences of two Nv1 isoforms with C-terminal Q (B1NWS1-1) and K (B1NWS1-2) are shown [https://www.uniprot.org/uniprot/B1NWS1] [138]. C-terminal residues of both isoforms are underlined. The disulfide bridges are shown above the alignment. The sequence similarity is shown as a dark (high) and light (low) gray background, the multiple sequence alignment was performed using the Vector NTI software.