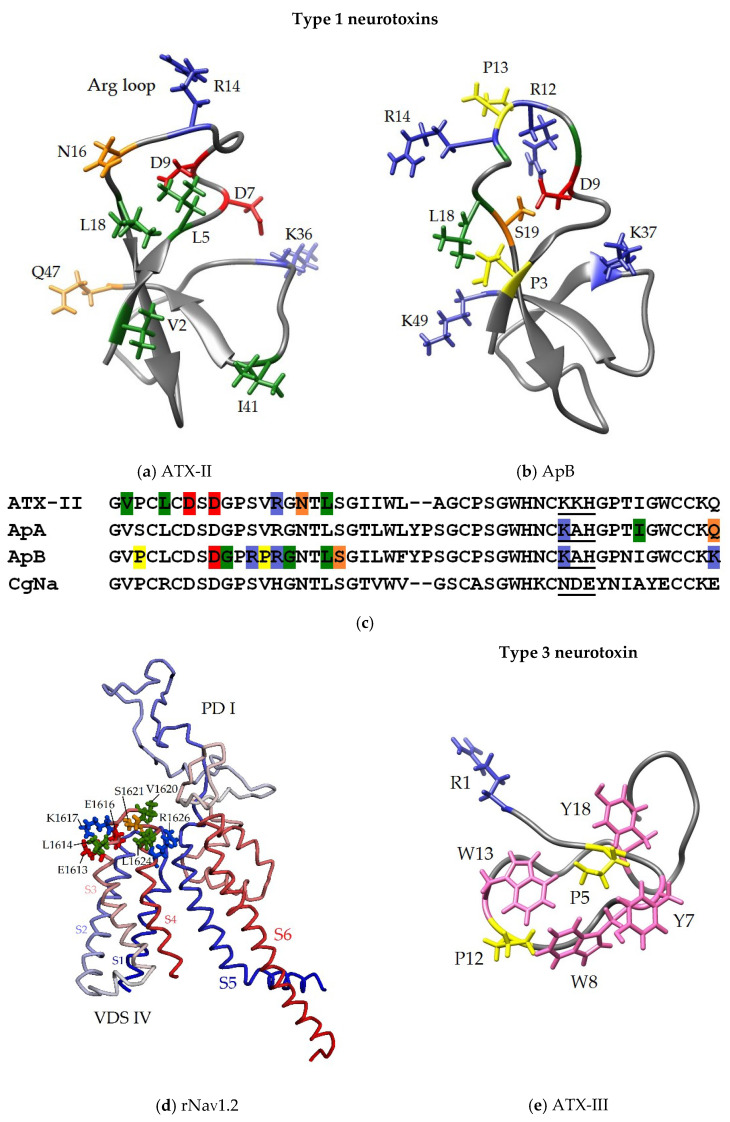
Figure 8.
Functionally significant amino acid residues of NaTxs (a–c,e) and ATX-II putative binding site (d). The homology models of ATX-II (based on ApA, PDB ID: 1Ahl) (a), and PD-I with VSD-IV of rNaV1.2 channel (based on hNaV1.2, PDB ID: 6J8E) (d), and 3D structure of ApB (1Apf) (b), and ATX-III (1ANS) (e). The ribbon diagram of the neurotoxins is shown, key residues presented as sticks and colored blue (basic), red (acidic), green (aliphatic), orange (polar), yellow (proline) and pink (aromatic). The ribbon diagram of rNaV1.2 fragment is colored from N- (blue) to C-termini (red), key ATX-II interacting residues are colored as described above. (c) Multiple alignment of the amino acid sequences of the type 1 NaTxs; key residues are colored as described above, and positively (ATX-III, ApA, ApB) or negatively (CgNa) charged motif is underlined.