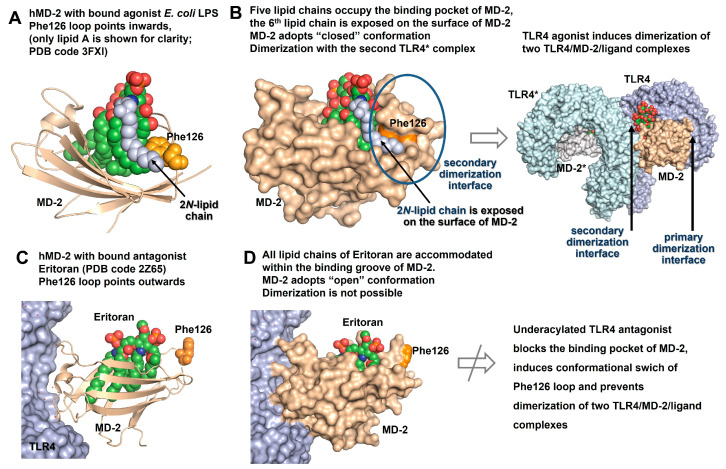
Figure 4.
Comparison of co-crystal structures of TLR4/MD-2-bound agonist and antagonist LPS/lipid A variants. (A) Co-crystal structure of hTLR4/MD-2/Ra-LPS (E. coli) complex (PDB code: 3FXI, only hMD-2 with bound lipid A portion of LPS is shown for clarity). The exterior positioning of the 2N-acyl chain (colored blue) of hTLR4/MD-2-bound lipid A is supported by interaction with Phe126 (colored orange) which points inwards due to ligand-driven conformational rearrangement in hMD-2. (B) Five lipid chains of hexa-acylated lipid A are accommodated in the binding pocket of hMD-2. The 6th 2N-linked hydroxyacyl residue is excluded from the binding pocket and, together with a side chain of Phe126, contributes to the formation of a secondary dimerization interface. The LPS binding induces dimerization of two TLR4/MD-2/LPS complexes. (C) All lipid chains of a tetra-lipidated TLR4 antagonist Eritoran are intercalated into the binding groove of hMD-2 (PDB code: 2Z65). Phe126 (colored orange) is exposed to solvent. (D) hMD-2 with bound antagonist ligand adopts an “open” conformation that results in prevention of dimerization. Images were generated with PyMol.