Abstract
The phylogenetic distributions of multiple putative virulence factors (VFs) and papA (P fimbrial structural subunit) alleles among 182 Escherichia coli blood isolates from patients with diverse-source bacteremia were defined. Phylogenetic correspondence among these strains, the E. coli Reference (ECOR) collection, and other collections of extraintestinal pathogenic E. coli (ExPEC) was assessed. Although among the 182 bacteremia isolates phylogenetic group B2 predominated, exhibited the greatest concentration of individual VFs, and contained the largest number of familiar virulent clones, other phylogenetic groups exhibited greater concentrations of certain VFs than did group B2 and included several additional virulent clones. Certain of the newly detected VF genes, e.g., fyuA (yersiniabactin; 76%) and focG (F1C fimbriae; 25%), were as prevalent or more prevalent than their more familiar traditional counterparts, e.g., iut (aerobactin; 57%) and sfaS (S fimbriae; 14%), thus possibly offering additional useful targets for preventive interventions. Considerable diversity of VF profiles was observed at every level within the phylogenetic tree, including even within individual lineages. This suggested that many different pathways can lead to extraintestinal virulence in E. coli and that the evolution of ExPEC, which involves extensive horizontal transmission of VFs and continuous remodeling of pathogenicity-associated islands, is a highly active, ongoing process.
The strains of Escherichia coli that cause extraintestinal infections such as urinary tract infection (UTI), meningitis, and bacteremia are distinct both from most intestinal commensal E. coli types and from diarrheagenic E. coli types (13, 57, 65, 70). These specialized extraintestinal pathogenic E. coli (ExPEC) strains (65) are thought to derive primarily from E. coli phylogenetic group B2 (as defined within the E. coli Reference [ECOR] collection by multilocus enzyme electrophoresis [MLEE]) (20, 54) and to acquire their unique pathogenicity from their distinctive virulence factors (VFs) (4, 10, 59, 60).
Putative VFs of ExPEC include diverse adhesins, toxins, polysaccharide coatings (including capsules and lipopolysaccharides), siderophores, serum resistance mechanisms, and invasins (21, 33, 70). Such VFs help the organisms colonize host surfaces, avoid and/or subvert host defense mechanisms, injure and/or invade host cells and tissues, and incite a noxious inflammatory response, thereby giving rise to clinical disease (12, 21, 70). The VF genes of ExPEC are thought to be primarily inherited vertically within evolutionary lineages but also to be transferred horizontally between lineages, in some instances on plasmids or on “pathogenicity-associated islands” (PAIs), which are gene blocks that contain multiple contiguous VF genes (2, 10, 16, 18, 36, 50, 61). Better understandings of the prevalence and evolutionary origins of the VFs of ExPEC and of the distinctive “virulent clones” that make up the ExPEC population should hasten the development of the preventive measures that are sorely needed against these morbid and costly infections (42, 43, 47, 55, 72).
Recent methodological and epidemiological developments now permit the ready detection of a broad array of putative VFs of ExPEC (23, 27, 30, 33) and multiple F antigen-specific alleles of P fimbrial structural subunit gene papA (25, 32, 35), all of which are potential candidates for VF-specific preventive interventions such as vaccines (42, 43, 47, 55). We previously applied these developments to the analysis of a collection of urosepsis isolates from Seattle, Wash. (30, 33, 35). We undertook the present study to gain an understanding of the prevalence and phylogenetic distribution of such traits among diverse-source E. coli bacteremia isolates. Specifically, we analyzed a large collection of E. coli blood isolates from patients with diverse-source bacteremia within which phylogenetic relationships were previously established by MLEE and for which O:H serotypes and selected VF markers have been reported (22, 50). We sought to characterize this population with respect to the prevalence and phylogenetic distribution of a broad range of “newer” VFs and of the 12 recognized papA alleles and to correlate the phylogenetic structure of this population with that of the ECOR collection and other collections of ExPEC from the literature.
MATERIALS AND METHODS
Strains.
The study population consisted of 182 of 187 previously published diverse-source E. coli bacteremia isolates from hospitalized or community-dwelling adults (50). (Five of the original 187 isolates were unavailable for analysis.) These strains were previously characterized with respect to genotype for papHC (P fimbriae), papEFG and papG (P fimbrial adhesin) alleles, hly (hemolysin), sfa and/or foc (S and F1C fimbriae), iut (aerobactin system), bma (M fimbriae), nfa (nonfimbrial adhesins), and afa and/or dra (afimbrial and fimbrial Dr antigen-binding adhesins); pulsed-field gel electrophoresis type; mannose-resistant hemagglutination and hemolysin phenotypes; O:H serotype; ribotype; and MLEE type (22, 50). Members of the ECOR collection (54) were provided by Howard Ochman and the American Type Culture Collection (Manassas, Va.). Strains were stored in 20% glycerol at −70°C until use.
Virulence genotypes.
In the present study the 182 bacteremia isolates were tested for 15 putative VF genes of ExPEC for which they had not previously been tested. Detection was by dot blot hybridization under stringent conditions, as previously described (27, 28, 33). Probes were generated and digoxigenin labeled using primers as previously described (28, 33). The VF genes encompassed five categories: adhesin, toxin, siderophore, capsule, and miscellaneous genes. The adhesin genes investigated were papAH (P fimbrial structural subunit), sfaS (S fimbrial adhesin), focG (the putative F1C fimbrial adhesin), and iha (iron-regulated gene homologue adhesin; a gene encoding a recently described putative adhesin found in ExPEC and E. coli O157:H7) (30, 74). The siderophore genes studied were fyuA (yersiniabactin) and iroN (a novel putative catecholate siderophore). The toxin genes studied were cnf1 (cytotoxic necrotizing factor) and cdtB (cytolethal distending toxin). The capsule genes studied were kpsMIITII and kpsMIIITIII (group II and group III capsular polysaccharide synthesis, respectively). The miscellaneous VF genes were cvaC (colicin V; multifunctional serum resistance-associated plasmids), traT (serum resistance associated), ibeA (invasion of brain endothelium), and malX, a marker for a PAI from archetypal ExPEC strain CFT073 (18, 33, 36).
Strains also were tested by PCR for three VF gene regions, including kpsMK1TK1 (K1 capsule synthesis), kpsMIITII, and sfaS, using primers and PCR conditions as previously described (33). Strains that were positive for kpsMIITII by dot blot hybridization but not by PCR were considered to be K2 capsule positive (33).
Strains that were positive for any pap element were tested for 12 alleles of papA, corresponding to the 11 established P fimbrial F types (F7-1, F7-2, and F8 to F16) plus the recently discovered F48 papA variant, by using a multiplex PCR-based assay, as previously described (23, 35).
Sequence determination of novel papA.
The papA DNA sequence was determined as previously described (35) for the three strains which were positive for papAH by blotting and PCR but negative for a defined papA allele in the allele-specific papA assay. Predicted PapA peptides were aligned with reference PapA sequences for traditional F types F7-1, F7-2, and F8 to F16 plus the recently described F48 PapA variant (35) and PapA from strain 536 (25, 32) by using CLUSTAL-W (75). An unrooted tree was inferred from the resulting PapA similarity matrix according to the NJ method (66) by using the application MEGA (40).
Phylogenetic comparison to the ECOR collection.
To relate the known phylogenetic structure of the bacteremia isolates (as previously defined by MLEE) (50) to the known phylogenetic structure of the ECOR collection, a subset of the 182 strains representing each of the four major phylogenetic divisions of the bacteremia collection (i.e., clusters I, III, IV, and V) were categorized as to ECOR phylogenetic group based on random amplified polymorphic DNA (RAPD) analysis (3, 24, 26, 32, 34, 77). Fingerprints were generated in duplicate for each strain using (separately) arbitrary decamer primers 1247, 1254, and 1283 (3). Amplification conditions were as described previously (3) except that commercial PCR beads were used (3, 24, 26, 32, 34, 77). Each group of fingerprints from the three primers was combined in series to create a “virtual” composite fingerprint. Comparable fingerprints were generated in parallel for 10 ECOR control strains, including two from each of the five major ECOR phylogenetic groups as defined by Herzer et al., based on electrophoretic mobility polymorphisms (20). Each bacteremia isolate was inferred to derive from the phylogenetic group of the ECOR strain with which it exhibited the highest Pearson correlation coefficient in pair-wise comparisons based on analog densitometric scans of composite RAPD fingerprints (24, 26, 27, 32, 34).
Cluster analysis of VF profiles.
For a subset of the bacteremia strains (i.e., those in electrophoretic type [ET] 47), a “tree” based on extended VF profiles was constructed according to the unweighted pair group method with averaging (UPGMA) (71) by using the application NT-SYSpc (Exeter Software, Setauket, N.Y.).
Statistical methods.
Comparisons of proportions were tested using Fisher's exact test (two tailed). Comparisons of the prevalences of different traits within the same population were made using McNemar's test (15). A VF score was calculated for each strain as the sum of that for all VFs for which the strain was positive, with values for the four pap regions (papA, papHC, papEFG, and papG) and the three sfa and/or foc elements (sfa and/or foc, sfaS, and focG) proportionally adjusted to account for multiple detection of the same operon. Aggregate VF scores for groups were compared using the Mann-Whitney U test. Correlations between VFs were tested using the phi coefficient, a chi-square-based measure of association for 2 by 2 tables. Because of multiple comparison, the threshold for statistical significance was a P value of <0.01, with a P value of <0.05 considered to reflect borderline statistical significance.
Nucleotide sequence accession numbers.
Newly determined papA sequences for strains CA033, CA039, and BOS020 were deposited in GenBank under accession no. AF332518, AF332519, and AF332520, respectively.
RESULTS
Prevalence of VFs and papA alleles.
All of the newly assessed VFs were detected at least once in the population (Table 1). Among the adhesin genes, the recently described iha, although significantly less prevalent than papA or papG (P < 0.001; McNemar's test), was as prevalent as sfa and/or foc and was significantly more prevalent than the other mannose-resistant adhesin genes assessed, i.e., afa and/or dra, nfa, and bma (P < 0.001 for all comparisons; McNemar's test). focG was significantly more prevalent than the better-known sialosyl-binding adhesin gene sfaS (P = 0.008; McNemar's test). Although hly was the most prevalent toxin gene, cnf1 and diarrhea-associated cdtB also were appreciably prevalent. Among the siderophore genes, fyuA (yersiniabactin), which only recently has been shown to occur in E. coli (33, 67), was significantly more prevalent than iut (aerobactin; P < 0.001; McNemar's test), and the prevalence of recently described iroN approached that of iut. Nearly 80% of the population exhibited group II capsule synthesis genes (kpsMIITII), with the K1 and the K2 variants accounting for 14 and 25% of kpsMIITII-positive strains, respectively. Group III capsules were uncommon. Meningitis-associated ibeA occurred in 12% of strains. The PAI marker gene malX, present in 71% of strains, was the second most prevalent trait after kpsMIITII (Table 1).
TABLE 1.
Distribution of VF genes in relation to phylogenetic cluster among 182 E. coli bacteremia isolates
| Associated trait | No. (% of total) | Prevalence of associated trait (no. [%]) for phylogenetic group clustera:
|
|||
|---|---|---|---|---|---|
| I (n = 21) | III (n = 119) | IV (n = 19) | V (n = 22) | ||
| papA | 118 (65) | 3 (14)d | 96 (81)d | 3 (16)d | 16 (73) |
| F7-1 | 3 (2) | 0 | 2 (2) | 0 | 1 (5) |
| F7-2 | 14 (8) | 0 | 8 (7) | 0 | 6 (27)c |
| F8 | 7 (4) | 0 | 5 (4) | 0 | 2 (9) |
| F9 | 3 (2) | 0 | 2 (2) | 0 | 1 (5) |
| F10 | 45 (25) | 1 (5)b | 38 (32)c | 0c | 6 (27) |
| F11 | 17 (9) | 0 | 16 (13)c | 1 (5) | 0 |
| F12 | 5 (3) | 1 (5) | 3 (3) | 1 (5) | 0 |
| F13 | 9 (5) | 0 | 9 (8)b | 0 | 0 |
| F14 | 12 (7) | 0 | 12 (10)c | 0 | 0 |
| F15 | 3 (2) | 0 | 3 (3) | 0 | 0 |
| F16 | 9 (5) | 0 | 7 (6) | 0 | 2 (9) |
| F48 | 20 (11) | 1 (5) | 18 (15)b | 0 | 1 (5) |
| papG | 101 (55) | 1 (5)d | 87 (73)d | 2 (11)d | 11 (50) |
| Allele II | 58 (32) | 0d | 45 (38)b | 2 (11)b | 11 (50) |
| Allele III | 32 (18) | 0 | 31 (26)d | 0b | 0b |
| Allele II + III | 9 (5) | 0 | 9 (8)b | 0 | 0 |
| Allele I + III | 2 (1) | 0 | 2 (2) | 0 | 0 |
| sfa and/or foc | 66 (36) | 0d | 66 (55)d | 0d | 0c |
| sfaS | 25 (14) | 0 | 25 (21)d | 0 | 0d |
| focG | 45 (25) | 1 (5)b | 44 (37)d | 0c | 0c |
| afa and/or dra | 15 (8) | 6 (29)c | 6 (5)b | 0 | 3 (14) |
| iha | 69 (38) | 2 (10)c | 50 (42) | 2 (11)b | 15 (68)c |
| bma | 3 (2) | 1 (5) | 1 (1) | 0 | 1 (5) |
| nfa | 15 (8) | 2 (10) | 8 (7) | 4 (21) | 1 (5) |
| hly | 82 (45) | 0d | 75 (63)d | 0d | 7 (32) |
| cnf1 | 50 (27) | 1 (5)c | 49 (41)d | 0c | 0cf |
| cdtB | 9 (5) | 0 | 9 (8)b | 1 (5) | 0 |
| iroN | 83 (46) | 2 (10)d | 74 (62)d | 4 (21)b | 3 (14)c |
| fyuA | 139 (76) | 5 (24)d | 114 (96)d | 3 (14)d | 17 (77) |
| iut | 103 (57) | 11 (52) | 63 (53) | 11 (52) | 18 (82)b |
| kpsMIITII | 144 (79) | 10 (48)c | 107 (90)d | 7 (33)d | 19 (82) |
| K1 | 36 (20) | 2 (10) | 29 (24)b | 1 (5) | 4 (18) |
| K2e | 20 (11) | 2 (10) | 10 (8) | 0 | 8 (36)c |
| kpsMIIITIII | 9 (5) | 2 (10) | 6 (5) | 0 | 1 (5) |
| ibeA | 21 (12) | 1 (5) | 19 (16)b | 0 | 1 (5) |
| cvaC | 12 (7) | 0 | 10 (8) | 1 (5) | 1 (5) |
| traT | 52 (29) | 6 (29) | 31 (26) | 6 (29) | 9 (41) |
| PAI | 130 (71) | 2 (10)d | 113 (95)d | 3 (14)d | 12 (55) |
Data for cluster II not shown (n = 1). Data for papG alleles II and III. sfa and/or foc, afa and/or dra, hly, jut, nfa, and bma are as reported elsewhere (20, 45).
P < 0.05, indicated group versus all other strains.
P ≤ 0.01, indicated group versus all other strains.
P ≤ 0.001, indicated group versus all other strains.
Strains were considered K2 positive if positive for kpsMIITII by blot but not by PCR.
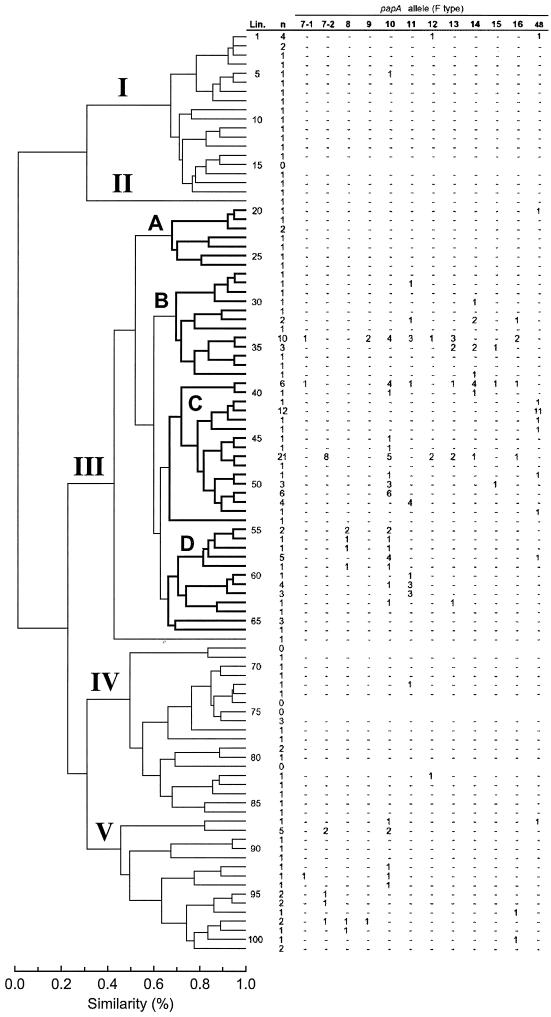
All 12 established papA alleles were detected at least once each in the population (Table 1). The recently described F48 papA variant (35) was the second most prevalent papA allele overall (n = 20), after the F10 variant (n = 45) (Table 1). Six strains were positive for papA by probe but negative for a defined papA allele in the F type-specific papA allele PCR assay. From five of these, full-length papA amplicons could be generated. Two of the papA amplicons repeatedly yielded uninterpretable sequence data; both were from strains known to have two copies of pap (not shown). Of the three amplicons that yielded interpretable sequence data, two proved to represent variants of the F11 papA allele. The third amplicon, from an isolate in ET 42, was distant from the 12 papA alleles included in the papA allele PCR assay but had 99.3% nucleotide identity with papA from archetypal pyelonephritis isolate 536 (25, 32).
Phylogenetic distribution of VFs.
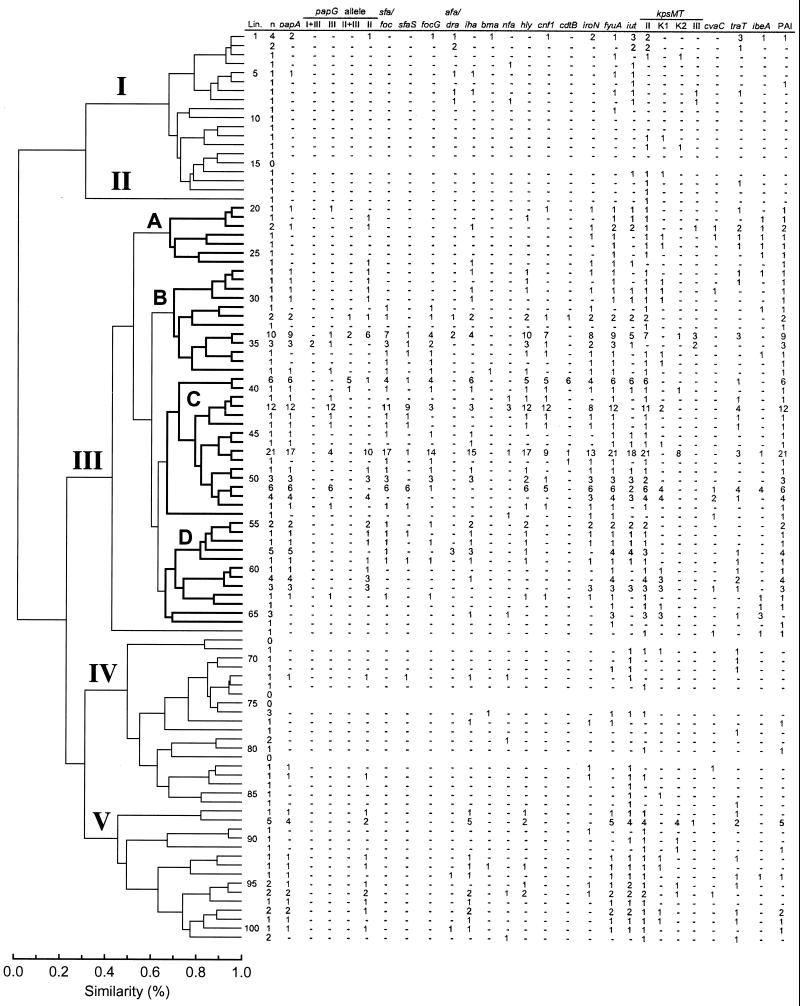
Projection of the VF and papA allele data onto the MLEE-based phylogenetic tree revealed diverse VF-specific patterns of distribution, which suggested both vertical and horizontal transmission of VFs, plus selective deletion of VFs from certain lineages (Fig. 1 and 2). Vertical inheritance of VFs within lineages was suggested by the significant concentration of specific VFs within particular phylogenetic groups, as was documented statistically at multiple hierarchical levels within the tree (Tables 1, 2, and 3). At the most basal level of the tree, i.e., that of major clusters (I to V), most VFs and papA alleles were significantly concentrated within the most populous cluster, cluster III. Exceptions included afa and/or dra (associated with cluster I) and the F7-2 papA allele, iha, iutA, and the K2 kpsMT variant (all associated with cluster V) (Table 1). Aggregate VF scores were highest in cluster III (median, 8.0; P < 0.001 versus all other strains and versus cluster I, IV, or V). They were intermediate in cluster V (median, 6.0; P > 0.10 versus all other strains and P < 0.001 versus cluster I or IV), and lowest in clusters I and IV (median, 2.0; P < 0.001 versus all other strains; P > 0.10 for cluster I versus cluster IV).
FIG. 1.
Phylogenetic distribution of VF genes among 182 E. coli bacteremia isolates. The MLEE-based tree was taken from Maslow et al. (50). Clusters I to V are labeled. The four major subclusters within cluster III, i.e., subclusters A to D, are in boldface. Data reflect the number of isolates within each lineage (Lin.) positive for the indicated trait. Minus signs, absence of positivity for the indicated trait; n, number of evaluatable isolates in the lineage.
FIG. 2.
Phylogenetic distribution of 12 F antigen-specific papA alleles (F types) among 182 E. coli bacteremia isolates. The MLEE-based tree is taken from Maslow et al. (50). Clusters I to V are labeled. The four major subclusters within cluster III, i.e., subclusters A to D, are in boldface. Data reflect the number of isolates within each lineage (Lin.) positive for the indicated papA allele. Minus signs, absence of positivity for the indicated allele; n, number of evaluatable isolates in the lineage. (In addition to the data shown, strain BOS020 from lineage 42 was positive for the F536 papA allele according to comparative sequence analysis.)
TABLE 2.
Phylogenetic distribution of VF genes within cluster III among 119 E. coli bacteremia isolatesa
| Associated trait | No. (% of 119) | Prevalence of associated trait (no. [%]) for subcluster:
|
|||
|---|---|---|---|---|---|
| A (n = 8) | B (n = 24) | C (n = 62) | D (n = 24) | ||
| papA | 96 (81) | 2 (25)d | 19 (79) | 56 (90)c | 19 (79) |
| F7-1 | 2 (2) | 0 | 1 (4) | 1 (2) | 0 |
| F7-2 | 8 (7) | 0 | 0 | 8 (13)c | 0 |
| F8 | 5 (4) | 0 | 0 | 0b | 5 (21)d |
| F9 | 2 (2) | 0 | 2 (8)b | 0 | 0 |
| F10 | 38 (32) | 0 | 5 (21) | 22 (35) | 11 (46) |
| F11 | 16 (14) | 0 | 4 (17) | 5 (8) | 7 (29)b |
| F12 | 3 (3) | 0 | 1 (4) | 2 (3) | 0 |
| F13 | 9 (8) | 0 | 5 (21)b | 3 (5) | 1 (4) |
| F14 | 12 (10) | 0 | 6 (25)b | 6 (10) | 0 |
| F15 | 3 (3) | 0 | 1 (4) | 2 (3) | 0 |
| F16 | 7 (6) | 0 | 5 (21)c | 2 (3) | 0 |
| F48 | 18 (15) | 1 (13) | 0b | 16 (26)d | 1 (4) |
| papG | 87 (73) | 4 (3) | 19 (16) | 51 (43)b | 13 (11)b |
| Allele II | 45 (38) | 3 (38) | 11 (46) | 19 (31) | 12 (50) |
| Allele III | 31 (26) | 1 (13) | 3 (13) | 26 (42)d | 1 (4)c |
| Allele II + III | 9 (8) | 0 | 3 (13) | 6 (10) | 0 |
| Allele I + III | 2 (2) | 0 | 2 (8)b | 0 | 0 |
| sfa and/or foc | 66 (56) | 0d | 15 (63) | 46 (74)d | 5 (21)d |
| sfaS | 25 (22) | 0 | 3 (13) | 20 (32)c | 2 (8) |
| focG | 44 (37) | 0b | 11 (46) | 29 (47)b | 4 (17)b |
| afa and/or dra | 6 (5) | 0 | 3 (8) | 0c | 3 (8) |
| iha | 50 (42) | 2 (25) | 9 (38) | 29 (47) | 10 (42) |
| bma | 1 (1) | 0 | 1 (4) | 0 | 0 |
| nfa | 8 (7) | 0 | 0 | 6 (10) | 2 (8) |
| hly | 75 (64) | 1 (13)c | 20 (83)b | 48 (77)d | 6 (25)d |
| cnf1 | 49 (42) | 1 (13) | 10 (42) | 37 (60)d | 1 (4)d |
| cdtB | 9 (8) | 0 | 1 (4) | 8 (13)b | 0 |
| iroN | 74 (63) | 4 (50) | 18 (75) | 44 (71) | 8 (33)c |
| fyuA | 114 (97) | 8 (100) | 21 (88) | 62 (100)b | 23 (96) |
| iut | 63 (53) | 5 (63) | 10 (42) | 36 (58) | 12 (50) |
| kpsMIITII | 107 (90) | 6 (75) | 18 (75)b | 60 (97)b | 22 (92) |
| K1 | 29 (25) | 2 (25) | 5 (21) | 11 (18) | 11 (46)b |
| K2e | 10 (8) | 0 | 1 (4) | 9 (15)b | 0 |
| kpsMIIITIII | 6 (5) | 1 (13) | 5 (21)d | 0c | 0 |
| cvaC | 10 (9) | 2 (25) | 1 (4) | 5 (8) | 1 (4) |
| traT | 31 (26) | 5 (63)b | 5 (21) | 14 (23) | 7 (29) |
| ibeA | 19 (15) | 5 (63)c | 3 (13) | 5 (8)b | 5 (21) |
| PAI | 114 (96) | 8 (100) | 23 (96) | 62 (100)b | 20 (83)c |
Data for papG alleles, sfa and/or foc, afa and/or dra, hly, iut, bma, and nfa are reported elsewhere (20, 45). One strain within cluster III fell outside the four subclusters. Thus, subclusters A to D account for only 118 strains, whereas the total in cluster III is 119 strains.
P < 0.05, indicated group versus all other strains.
P ≤ 0.01, indicated group versus all other strains.
P ≤ 0.001, indicated group versus all other strains.
Strains were considered K2 positive if positive for kpsMIITII by blot but not PCR.
TABLE 3.
Phylogenetic distribution of VF genes within subcluster IIIC among 62 E. coli bacteremia isolatesa
| Associated trait | No. (% of 62) | Prevalence of associated trait (no. [%]) for lineages:
|
|||
|---|---|---|---|---|---|
| 39 and 40 (n = 7) | 41–44 (n = 15) | 45–48 (n = 24) | 49–53 (n = 15) | ||
| papA | 56 (90) | 7 (100) | 15 (100) | 19 (79)b | 15 (100) |
| F7-1 | 1 (2) | 1 (14) | 0 | 0 | 0 |
| F7-2 | 8 (13) | 0 | 0 | 8 (33)d | 0 |
| F10 | 22 (35) | 5 (71) | 0d | 7 (29) | 10 (67)c |
| F11 | 5 (9) | 1 (14) | 0 | 0 | 4 (27)c |
| F12 | 2 (3) | 0 | 0 | 2 (8) | 0 |
| F13 | 3 (5) | 1 (14) | 0 | 2 (8) | 0 |
| F14 | 6 (10) | 5 (71)d | 0 | 1 (4) | 0 |
| F15 | 2 (3) | 1 (14) | 0 | 0 | 1 (7) |
| F16 | 2 (3) | 1 (14) | 0 | 1 (4) | 0 |
| F48 | 16 (26) | 0 | 14 (93)d | 0d | 2 (13) |
| papG | 50 (81) | 7 (100) | 15 (100)b | 14 (58)d | 15 (100)b |
| Allele II | 19 (31) | 1 (14) | 0c | 10 (42) | 8 (53)b |
| Allele III | 26 (42) | 0b | 15 (100)d | 4 (17)c | 7 (47) |
| Allele II + III | 6 (10) | 6 (86)a | 0 | 0 | 0 |
| sfa and/or foc | 46 (74) | 5 (71) | 13 (87) | 17 (71) | 11 (73) |
| sfaS | 20 (32) | 1 (14) | 11 (73)d | 1 (4)d | 7 (47) |
| focG | 29 (47) | 5 (71) | 3 (33)b | 16 (67)b | 5 (33) |
| iha | 29 (47) | 7 (100)c | 1 (7)d | 17 (71)c | 4 (27) |
| nfa | 6 (10) | 0 | 4 (27)b | 1 (4) | 0 |
| hly | 48 (77) | 6 (86) | 15 (100)b | 17 (71) | 10 (67) |
| cnf1 | 37 (60) | 6 (86) | 15 (100)d | 9 (38)c | 7 (47) |
| cdtB | 8 (13) | 6 (86)d | 0 | 2 (8) | 0 |
| iroN | 44 (71) | 5 (71) | 10 (67) | 14 (58) | 14 (93)b |
| fyuA | 62 (100) | 7 (100) | 15 (100) | 24 (100) | 15 (100) |
| iut | 36 (58) | 7 (100)b | 0d | 20 (83)c | 9 (60) |
| kpsMIITII | 60 (97) | 7 (100) | 14 (93) | 24 (100) | 15 (100) |
| K1 | 11 (18) | 0 | 2 (13) | 1 (4)b | 8 (53)d |
| K2e | 9 (15) | 1 (14) | 0 | 8 (33)d | 0 |
| cvaC | 5 (8) | 0 | 0 | 0 | 4 (27)c |
| traT | 14 (23) | 1 (14) | 5 (33) | 3 (13) | 5 (33) |
| ibeA | 5 (8) | 0 | 0 | 1 (4) | 4 (27)c |
| PAI | 62 (100) | 7 (100) | 15 (100) | 24 (100) | 15 (100) |
Data for papG alleles, sfa and/or foc, afa and/or dra, hly, iut, bma, and nfa are as reported elsewhere (20, 45). One strain within subcluster C (lineage 54) fell outside the four major groups of lineages. Thus, lineages 39 to 53 account for only 61 strains, whereas the total in subcluster C is 62 strains.
P < 0.05, indicated group versus all other strains.
P ≤ 0.01, indicated group versus all other strains.
P ≤ 0.001, indicated group versus all other strains.
Strains were considered K2 positive if positive for kpsMIITII by blot but not PCR.
A nonrandom phylogenetic distribution of VFs was apparent also at the level of the major subclusters (A to D) within cluster III (Table 2). Here, however, no consistent pattern as to which subcluster exhibited the greatest prevalence of specific traits emerged (Table 2). Similar findings were also made at the level of the four groups of individual lineages that made up subcluster C, the most populous subcluster within cluster III (Table 3).
Exceptionally, particular VFs or papA alleles exhibited an “all or none” pattern of phylogenetic distribution, either occurring in all members of a phylogenetic group or being entirely absent from the group. Most traits, however, if present at all occurred in only a fraction of the members of any particular phylogenetic group, even if that group exhibited a significantly higher prevalence of the trait than did comparable groups at the same hierarchical level in the tree (Fig. 1 and 2; Tables 1 to 3). This phenomenon could be explained by selective gene deletion from members of an ancestrally VF-positive group and/or by horizontal entry of the trait into multiple smaller branches of the tree in the absence of a VF-positive common ancestor. Strongly favoring horizontal transfer in some instances was the markedly discontinuous distribution of certain VFs across the tree, with many intervening VF-negative lineages, a distribution pattern that would require multiple deletion events if not due to horizontal transfer (Fig. 1 and 2; Tables 1 to 3). Even more compelling evidence of horizontal transfer was provided by the distribution of papA alleles, since adjacent lineages commonly exhibited different alleles, whereas each shared its particular allele with phylogenetically distant lineages (Fig. 2; Table 3). Moreover, many individual lineages exhibited diverse papA alleles among their various members. These phenomena could not be explained by selective deletion alone and instead suggested horizontal exchange of papA alleles between lineages, possibly even independent of other pap elements.
Ordered diversity of VF genotypes within an individual lineage.
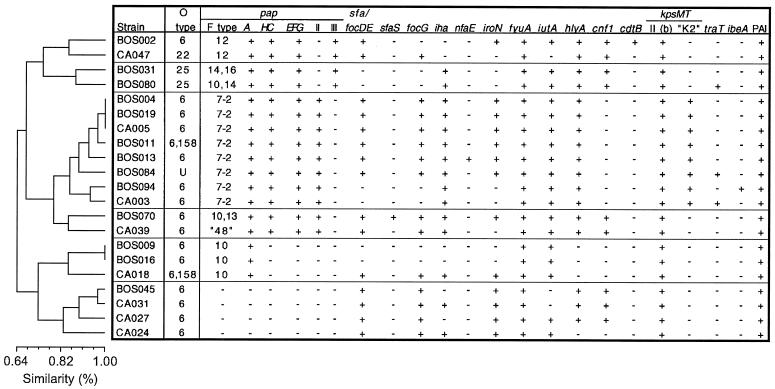
Because of limiting numbers, statistical comparisons of aggregate VF data (e.g., Tables 1 to 3) could not be made at the level of the individual lineages (i.e., ETs) within the lineage groups shown in Table 3, although diversity of VF profiles was still apparent even at this level (Fig. 1 and 2). However, ET 47, the single most populous lineage overall, provided an opportunity to analyze the continuing diversity of VF profiles even at the most differentiated extreme of the phylogenetic tree. The members of ET 47 exhibited seven distinct papA alleles and two papG allele configurations and were variably positive for most of the other traits detected within this lineage, with only fyuA, kpsMIITII, and the PAI marker being universally prevalent (Fig. 3). Analysis of the VF and papA allele data for ET 47 by UPGMA revealed that these traits were not randomly distributed (Fig. 3). Instead, they segregated in an arborizing pattern that could be explained most parsimoniously by the sequential acquisition or loss of specific markers by separate subclones within this lineage as the subclones progressively differentiated from a common ancestor (Fig. 3).
FIG. 3.
Cluster analysis of VF and papA allele data for ET 47. Dendrogram (according to UPGMA) reflects similarity relationships based on VF and papA allele profiles among isolates from ET 47, which have indistinguishable genomic backgrounds according to MLEE. Strain CA039 was negative in the F PCR assay but contained a papA variant most similar to the F48 papA allele according to comparative sequence analysis.
Associations between individual VFs.
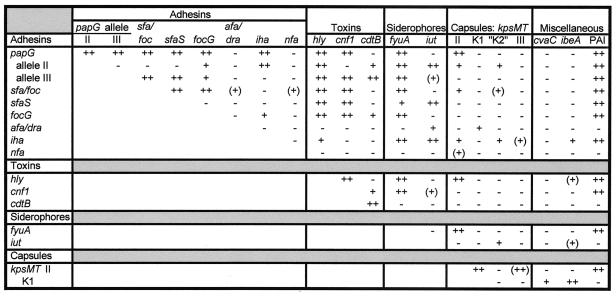
Analysis of the total VF data set revealed many familiar patterns of association between VFs (23, 30, 33, 35). These included the positive associations among “J96-like” traits sfa and/or foc, hly, cnf1, and papG allele III, as a group, and among “CFT073-like” traits iha, iut, papG allele II, kpsMIITII, and the K2 kpsMT variant, as a group, together with the generally negative associations between these two groups (Fig. 4). Also of note were the negative associations of afa and/or dra and nfa with sfa and/or foc, the association of ibeA with the (meningitis-associated) K1 kpsMT variant, and the absence of an association between iha and the PAI marker gene malX despite their close physical proximity on the same PAI in archetypal strain CFT073 (18, 36).
FIG. 4.
Correlations among VF genes for 182 E. coli bacteremia isolates. Only those VF genes that yielded at least one correlation at the P ≤ 0.01 level are shown. Results for papA were similar to those for papG. Significance levels: −, P > 0.01; +, P ≤ 0.01; ++, P ≤ 0.001. (Parentheses denote negative associations.)
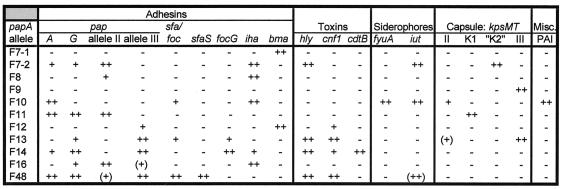
The various papA alleles also exhibited multiple distinctive associations with specific VF genes (Fig. 5). Of note were the nonoverlapping groups of papA alleles associated with certain closely related VF genes such as papG allele II versus allele III, sfaS versus focG, and the various kpsMT variants (Fig. 5). Also of note were the shared associations of hly and cnf1 with the F13, F14, and F48 papA alleles but the specific associations of hly with the F7-2 papA allele and of cnf1 with the F12 papA allele (Fig. 5).
FIG. 5.
Correlations among F antigen-specific papA alleles and other VF genes for 182 E. coli bacteremia isolates. Only those papA alleles that yielded at least one correlation at the P ≤ 0.01 level are shown. Significance levels: −, P > 0.01; +, P ≤ 0.01; ++, P ≤ 0.001. (Parentheses denote negative associations.)
Comparisons among the papA alleles themselves revealed associations between the F8 allele and both the F9 and F10 alleles (P < 0.01), between the F14 and F16 alleles (P < 0.001), and between the F9 and F13 alleles (P < 0.001). Many additional associations were noted at the level of P < 0.05 (not shown).
Correspondence of the present population with ECOR groups and with known virulent clones.
Composite RAPD analysis revealed that clusters I, III, IV, and V of the present population (Fig. 1 and 2) correspond, respectively, with ECOR phylogenetic groups A, B2, B1, and D, as defined by MLEE (not shown). These findings, together with previously determined serotypes for the present population (50), the VF and papA data from the present study, and selective additional comparative RAPD analyses (not shown), allowed certain lineages to be presumptively linked with recognized virulent clones of ExPEC (Table 4). Of note, two of the distinct pathotypic groups within ET 47 (Fig. 3), one characterized by the F12 papA allele, papG allele III, and both hly and cnf and the other characterized by the F7-2 papA allele, papG allele II, and hly but not cnf, corresponded to two recognized virulence-associated clones within serogroup O6 (Table 4). Because of the multiple VF profile subdivisions within ET 47 (Fig. 3), ET 42, which was the second most populous lineage overall but which in comparison with ET 47 was pathotypically fairly homogeneous (Fig. 1 and 2), emerged as the most prevalent clonopathotype (not shown). This ET, one of whose members (BOS020) exhibited the recently described “F536” papA variant, putatively corresponds to an O6:K+;F48/F536-positive lineage, which is a prominent cause of UTI in dogs, cats, and humans and which includes archetypal pyelonephritis isolate 536 (Table 4).
TABLE 4.
Proposed correspondence between specific lineages from present study and recognized virulent clones of ExPEC
| ET lineage | Putative clonal group | Corresponding clonal groups from literature
|
Comments | ||||||
|---|---|---|---|---|---|---|---|---|---|
| ECOR (23, 54) | Selander et al. (69) | Ochman and Selander (53) | Achtman et al. (1) pattern(s) | Cherifi et al. (11) clone | Whittam et al. (25, 80) ET | Vaisanen-Rhen et al. (76) clone(s) | |||
| 29, 30 | O16:K1 | j,k/12 | 12 | VI | Pyelonephritis, sepsis | ||||
| 34, 35 | O4:K3/10:H5 | 59, 60 | ET 18 | 12 and 14 | IV | Archetypal strains I96 and CP9 (29, 31) | |||
| 39 | O2:K5/7:H1 | 57 | Urosepsis; usually cdt positive (33) | ||||||
| 42 | O6:K15/53:H31 | 2 | 21 | Archetypal strain 536; UTI in dogs and cats; cystitis in humans (25, 27, 32, 34) | |||||
| 47 | O6:K2:H1 | 56 | Cluster B | 1 | 1 | V | Archetypal strain CFT073 (35, 52) | ||
| 47 | O6:K13/K53:H1 | 56 | Cluster B | 1 | 1 | VIII | Humans and pets; cdt positive or negative (25, 34) | ||
| 51 | O18:K1:H7 | (61, 62)a | Cluster A | f,g,h,i/6,9 | 6 and 9 | 4 and 6 | I and II | Archetypal neonatal meningitis isolates RS218 and C5; cystitis in women (9, 24) | |
| 52, 61-62 | O1/O2:K1:H7 | 61, 62 | Cluster A | e/6,9 | 6 and 9 | 5 and 6 | I and II | Meningitis and sepsis, pyelonephritis | |
| 55, 56 | O18:K5:H−/H7 | Cluster E | 11 | VII | UTI | ||||
| 58, 59 | O75:K5/100:H5 | 64 | 11 | Usually afa and/or dra positive (39) | |||||
| 93 | O7:K1:H− | 38–41 | Cluster H | 1/3,15,16,m/3 | 3 | Neonatal meningitis and sepsis | |||
| 95–97 | O15:K52:H1 | (44)a | Epidemic and endemic UTI (56, 58, 62) | ||||||
| 98–99 | O1:K1:H− | 35, 36 | a/5 | 5 | Neonatal meningitis and sepsis | ||||
| 100 | O157:K+:H+ | UTI, urosepsis (33, 78) | |||||||
Correspondence with indicated ECOR strains is approximate.
DISCUSSION
In the present study we assessed the phylogenetic distribution of multiple VF genes and papA alleles among E. coli isolates from patients with diverse-source bacteremia and defined the phylogenetic correspondence between these strains, the ECOR collection, and other collections of ExPEC. We found that although among the bacteremia isolates phylogenetic group B2 predominated overall, other phylogenetic groups exhibited greater concentrations of certain VFs than did group B2 and included several additional recognized virulent clones. Moreover, certain of the newly detected VFs were as prevalent or more prevalent than their more familiar traditional counterparts, thus possibly constituting additional useful targets for preventive interventions. The considerable diversity of VF profiles observed at every level within the phylogenetic tree, including even within individual lineages, suggested that many different pathways can lead to extraintestinal virulence in E. coli and that the evolution of ExPEC, which involves extensive horizontal transmission of VFs and continuous remodeling of PAIs, is an active, ongoing process.
Our results are consistent with previous evidence that E. coli phylogenetic group B2 predominates among ExPEC and is the main repository (hence, presumably, the original source within the species) of many extraintestinal VFs (10, 23, 33, 45, 59, 60). Our data considerably enlarge the range of VFs known to be associated with phylogenetic group B2 in the context of diverse-source bacteremia (22, 50). However, they also demonstrate that certain VFs are significantly concentrated outside of group B2 in phylogenetic group A or D or are broadly distributed in the population without a focal concentration in any one phylogenetic group. This is consistent with recent findings derived from analysis of the ECOR collection and urosepsis isolates (23, 30, 33). The latter observations suggest that the evolutionary histories of the various VFs of ExPEC are more complex and diverse than can be accounted for by two-step models that focus primarily on phylogenetic group B2, as previously proposed based on an analysis of pap, hly, sfa and/or foc, and kps (10, 45).
The broad range of our VF gene screening allowed us to document an appreciable prevalence and/or nonrandom phylogenetic distribution for many newer VF genes, including iha, fyuA, iroN, cnf, cdt, and ibeA. Likewise, by discriminating between closely related variants of a particular VF gene or gene family we were able to detect disparate associations, prevalences, and phylogenetic backgrounds for the 12 papA alleles, sfaS versus focG, and the several kpsMT variants. This simultaneously broad and highly discriminating approach to VF genotyping provided the added advantage of generating distinctive extended VF “signatures” for individual strains and lineages. In many instances, with or without supplemental data these signatures allowed us to associate lineages from the present population with known virulent clones from the literature (Table 4).
For example, we found that an O6:K+;F48/F536 clonal group that is associated with UTI in animals and humans (unpublished data; 25, 27, 34) and that includes archetypal pyelonephritis isolate 536 (8) was the single most prevalent clonopathotype in the study population. It accounted for ∼7% of isolates overall, and its isolates outnumbered even those of the prominent O6:K2:H1 clonal group (∼4%). The associated primary sources of bacteremia for these O6:K+;F48/F536 strains included UTI, pneumonitis, and intra-abdominal infections (49). The evident pathogenetic versatility of this clone with respect to host species, clinical syndrome, and anatomical site of infection illustrates the inappropriateness of the traditional designation of uropathogenic E. coli and supports instead a more inclusive rubric such as ExPEC (65). Consistent with this “generalist” concept, we identified within the study population several representatives of the O18:K1:H7 clonal group, which traditionally has been associated with neonatal meningitis (9, 38, 69) but which recently was shown to be a major contributor also to uncomplicated cystitis in women (24, 26, 41).
Correlation of the newly detected VFs, and especially the papA alleles, with the population's underlying phylogenetic structure suggested even more extensive horizontal transfer of VFs than was previously proposed for this population based on analysis of pap, hly, sfa and/or foc, afa and/or dra, and iut (50) or the three papG alleles (22). The evidence of horizontal transfer extended down to the most differentiated subdivisions of the phylogenetic tree, i.e., the individual ETs themselves, and beyond. This suggests that ExPEC strains are actively evolving with respect to VF profiles even today and hence represent a “moving target” for the pathotypic analyses that are needed to guide the development of VF-specific protective measures. Moreover, the ordered diversity of VF profiles observed within ET 47 (Fig. 3) suggests ongoing evolution of the genomic background at a level below the detection threshold of MLEE, which is insensitive to silent mutations and to peptide polymorphisms that do not alter the electrophoretic mobility of the particular markers analyzed (68, 79). We propose that in the instance of ET 47 the VF data, rather than being phylogenetically unreliable (as was previously suggested when diverse pathotypes were encountered within a single electrophoretic lineage) (69), actually supersede MLEE by revealing significant evolutionary diversity that MLEE is unable to detect. Formal testing of this hypothesis should be possible with the use of a phylotyping method more discriminating than MLEE, e.g., multilocus sequence typing (14, 45, 48, 63).
Although the strict clinical criteria for inclusion in the present study population (50) represent an epidemiological strength of the study, they do limit the generalizability of the findings to the larger E. coli population. The requirement that isolates must have caused bacteremia imposes a substantial “filter” that is predicted to bias the population not only toward the more virulent phylogenetic groups such as B2 and D (i.e., clusters III and V) but also toward the more virulent members within each group. This filtering effect would probably be strongest for intrinsically “low-virulence” groups such as A and B1 (i.e., clusters I and IV), most members of which may lack VFs and hence would be unable to cause bacteremia except in the presence of significant host compromise (23, 60). Consistent with this hypothesis, multiple VFs were numerically more prevalent within cluster I of the present population (group A equivalent) than among the group A ECOR strains (23). The genes for these included afa and/or dra (29 versus 0%), iut (52 versus 24%), kpsMIITII (48 versus 20%), traT (29 versus 16%), and nfa, kpsIIIMTIII, and malX (all 10 versus 0%). In contrast, only two VF genes were more prevalent among the group A ECOR strains, i.e., iha (28 versus 10%) and fyuA (52 versus 24%) (23). Similar trends were evident when cluster IV of the present population (group B1 equivalent) was compared with the group B1 ECOR strains (not shown) (23). Thus, broad phylogenetic conclusions must be tempered by recognition of the likely substantial influence of clinical context on the observed associations.
Previous studies have documented that PAIs from different ExPEC strains can differ considerably with respect to their constituent VF genes (9, 18, 19, 64, 73) and, for those VF genes that do consistently occur together in multiple PAIs, the specific genetic linkages between the VF genes (46). These largely anecdotal observations, together with our present population-based finding that even VFs known to be PAI associated in certain strains commonly exhibit divergent patterns of phylogenetic distribution both among lineages and even within a given lineage, strongly suggest that VF genes are highly mobile independent of PAIs and that PAIs are subject to continuous, ongoing remodeling. These data are consistent with a model in which PAIs, although perhaps occasionally subject to en bloc horizontal transfer (10, 16) or total deletion (5, 8, 19, 37), participate to a much greater extent in the horizontal transfer of VFs by providing genomic regions receptive to the insertion, retention, and release of individual VF genes.
The abundance of recombination-promoting elements within PAIs (16) presumably allows for the ready acquisition of any “wandering” VF genes that happen to enter the cell, plus ready deletion of previously integrated VF genes, including their donation to nearby plasmids, transposons, etc. The resulting rapid reassortment of VF genes within PAIs continuously generates novel combinations of VFs on which selective pressures can act. Those combinations of VFs that increase fitness in a particular niche or that broaden the range of niches in which the host cell can effectively compete are conserved and expand with the clone. However, just as the clonal frame of the bacterial host's (presumably selection-neutral) genomic background gradually breaks down over time from recombined genomic regions (17, 44, 51), so too is the host's highly selectable VF profile (i.e., the architecture of its PAIs) disrupted by newly acquired and deleted VF genes. According to this model, PAIs are not so much horizontally mobile and internally stable as they are horizontally stable but internally highly dynamic. Long-range VF mapping (7), comparative genomic analysis (6), and long-term culture experiments involving clonally mixed populations may allow direct testing of this model in the future.
In summary, we found that among E. coli blood isolates from patients with diverse-source bacteremia, phylogenetic group B2 predominated overall and accounted for most of the individual VFs and virulent clones. However, other phylogenetic groups exhibited greater concentrations of certain VFs than did group B2 and accounted for additional virulent clones. Certain newly detected VFs were as prevalent or more prevalent than were their more familiar counterparts and hence may constitute useful targets for future preventive interventions. The considerable diversity of VF profiles noted at every level within the phylogenetic tree suggests that many different pathways can lead to extraintestinal virulence in E. coli and that the evolution of ExPEC, which involves extensive horizontal transmission of VFs and continuous remodeling of PAIs, is a highly active and ongoing process.
ACKNOWLEDGMENTS
This work was supported by the Office of Research and Development, Medical Research Service, Department of Veterans Affairs (J.R.J. and J.N.M.), National Institutes of Health grant DK-47504 (J.R.J), and National Research Initiative (NRI) Competitive Grants Program/United States Department of Agriculture grant 00-35212-9408 (J.R.J.).
Dave Prentiss (Minneapolis VAMC) helped prepare the figures. Ann Emery (Minneapolis VAMC) helped with manuscript preparation.
REFERENCES
- 1.Achtman M, Mercer A, Kusecek B, Pohl A, Heuzenroeder M, Aaronson W, Sutton A, Silver R P. Six widespread bacterial clones among Escherichia coli K1 isolates. Infect Immun. 1983;39:315–335. doi: 10.1128/iai.39.1.315-335.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Arthur M, Arbeit R D, Kim C, Beltran P, Crowe H, Steinback S, Campanelli C, Wilson R A, Selander R K, Goldstein R. Restriction fragment length polymorphisms among uropathogenic Escherichia coli isolates: pap-related sequences compared with rrn operons. Infect Immun. 1990;58:471–479. doi: 10.1128/iai.58.2.471-479.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Berg D E, Akopyants N S, Kersulyte D. Fingerprinting microbial genomes using the RAPD or AP-PCR method. Methods Mol Cell Biol. 1994;5:13–24. [Google Scholar]
- 4.Bingen E, Picard B, Brahimi N, Mathy S, Desjardins P, Elion J, Denamur E. Phylogenetic analysis of Escherichia coli strains causing neonatal meningitis suggests horizontal gene transfer from a predominant pool of highly virulent B2 group strains. J Infect Dis. 1998;177:642–650. doi: 10.1086/514217. [DOI] [PubMed] [Google Scholar]
- 5.Bloch C A, Rode C K. Pathogenicity island evaluation in Escherichia coli K1 by crossing with laboratory strain K-12. Infect Immun. 1996;64:3218–3223. doi: 10.1128/iai.64.8.3218-3223.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bloch C A, Rode C K, Obreque V, Russell K Y. Comparative genome mapping with mobile physical map landmarks. J Bacteriol. 1994;176:7121–7125. doi: 10.1128/jb.176.22.7121-7125.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Blum G, Ott M, Cross A, Hacker J. Virulence determinants of Escherichia coli O6 extraintestinal isolates analysed by Southern hybridizations and DNA long range mapping techniques. Microb Pathog. 1991;10:127–136. doi: 10.1016/0882-4010(91)90073-j. [DOI] [PubMed] [Google Scholar]
- 8.Blum G, Ott M, Lischewski A, Ritter A, Imrich H, Tschape H, Hacker J. Excision of large DNA regions termed pathogenicity islands from tRNA-specific loci in the chromosome of an Escherichia coli wild-type pathogen. Infect Immun. 1994;62:606–614. doi: 10.1128/iai.62.2.606-614.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bonacorsi S P P, Clermont O, Tinsley C, Le Gall I, Beaudoin J-C, Elion J, Nassif X, Bingen E. Identification of regions of the Escherichia coli chromosome specific for neonatal meningitis-associated strains. Infect Immun. 2000;68:2096–2101. doi: 10.1128/iai.68.4.2096-2101.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Boyd E F, Hartl D L. Chromosomal regions specific to pathogenic isolates of Escherichia coli have a phylogenetically clustered distribution. J Bacteriol. 1998;180:1159–1165. doi: 10.1128/jb.180.5.1159-1165.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cherifi A, Contrepois M, Picard B, Oullet P, Orskov I, Orskov F, De Rycke J. Clonal relationships among Escherichia coli serogroup O6 isolates from human and animal infections. FEMS Microbiol Lett. 1991;80:225–230. doi: 10.1016/0378-1097(91)90600-f. [DOI] [PubMed] [Google Scholar]
- 12.Donnenberg M S, Welch R A. Virulence determinants of uropathogenic Escherichia coli. In: Mobley H L T, Warren J W, editors. Urinary tract infections: molecular pathogenesis and clinical management. Washington, D.C.: ASM Press; 1996. pp. 135–174. [Google Scholar]
- 13.Eisenstein B I, Jones G W. The spectrum of infections and pathogenic mechanisms of Escherichia coli. Adv Intern Med. 1988;33:231–252. [PubMed] [Google Scholar]
- 14.Enright M C, Spratt B C. Multilocus sequence typing. Trends Microbiol. 1999;7:482–487. doi: 10.1016/s0966-842x(99)01609-1. [DOI] [PubMed] [Google Scholar]
- 15.Fleiss J L. Statistical methods for rates and proportions. New York, N.Y: John Wiley & Sons; 1981. pp. 112–137. [Google Scholar]
- 16.Groisman E A, Ochman H. Pathogenicity islands: bacterial evolution in quantum leaps. Cell. 1996;87:791–794. doi: 10.1016/s0092-8674(00)81985-6. [DOI] [PubMed] [Google Scholar]
- 17.Guttman D S, Dykhuizen D E. Clonal divergence in Escherichia coli as a result of recombination, not mutation. Science. 1994;266:1380–1383. doi: 10.1126/science.7973728. [DOI] [PubMed] [Google Scholar]
- 18.Guyer D M, Kao J-S, Mobley H L T. Genomic analysis of a pathogenicity island in uropathogenic Escherichia coli CFT073: distribution of homologous sequences among isolates from patients with pyelonephritis, cystitis, and catheter-associated bacteriuria and from fecal samples. Infect Immun. 1998;66:4411–4417. doi: 10.1128/iai.66.9.4411-4417.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hacker J, Bender L, Ott M, Wingender J, Lund B, Marre R, Goebel W. Deletions of chromosomal regions coding for fimbriae and hemolysins occur in vitro and in vivo in various extraintestinal Escherichia coli isolates. Microb Pathog. 1990;8:213–225. doi: 10.1016/0882-4010(90)90048-u. [DOI] [PubMed] [Google Scholar]
- 20.Herzer P J, Inouye S, Inouye M, Whittam T S. Phylogenetic distribution of branched RNS-linked multicopy single-stranded DNA among natural isolates of Escherichia coli. J Bacteriol. 1990;172:6175–6181. doi: 10.1128/jb.172.11.6175-6181.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Johnson J R. Virulence factors in Escherichia coli urinary tract infection. Clin Microbiol Rev. 1991;4:80–128. doi: 10.1128/cmr.4.1.80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Johnson J R, Brown J J, Maslow J N. Clonal distribution of the three alleles of the Gal(α1–4)Gal-specific adhesin gene papG among Escherichia coli strains from patients with bacteremia. J Infect Dis. 1998;177:651–661. doi: 10.1086/514230. [DOI] [PubMed] [Google Scholar]
- 23.Johnson J R, Delavari P, Kuskowski M, Stell A L. Phylogenetic distribution of extraintestinal virulence-associated traits in Escherichia coli. J Infect Dis. 2001;183:78–88. doi: 10.1086/317656. [DOI] [PubMed] [Google Scholar]
- 24.Johnson J R, Delavari P, O'Bryan T. Escherichia coli O18:K1:H7 isolates from acute cystitis and neonatal meningitis exhibit common phylogenetic origins and virulence factor profiles. J Infect Dis. 2001;183:425–434. doi: 10.1086/318086. [DOI] [PubMed] [Google Scholar]
- 25.Johnson J R, Delavari P, Stell A L, Whittam T S, Carlino U, Russo T A. Molecular comparison of extraintestinal Escherichia coli isolates from the same electrophoretic lineages from humans and domestic animals. J Infect Dis. 2001;183:154–159. doi: 10.1086/317662. [DOI] [PubMed] [Google Scholar]
- 26.Johnson J R, O'Bryan T T, Delavari P, Kuskowski M, Stapleton A, Carlino U, Russo T. Clonal relationships and extended virulence genotypes among Escherichia coli isolates from women with a first or recurrent episode of cystitis. J Infect Dis. 2001;183:1508–1517. doi: 10.1086/320198. [DOI] [PubMed] [Google Scholar]
- 27.Johnson J R, O'Bryan T T, Low D A, Ling G, Delavari P, Fasching C, Russo T A, Carlino U, Stell A L. Evidence of commonality between canine and human extraintestinal pathogenic Escherichia coli strains that express papG allele III. Infect Immun. 2000;68:3327–3336. doi: 10.1128/iai.68.6.3327-3336.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Johnson J R, Russo T A, Brown J J, Stapleton A. papG alleles of Escherichia coli strains causing first episode or recurrent acute cystitis in adult women. J Infect Dis. 1998;177:97–101. doi: 10.1086/513824. [DOI] [PubMed] [Google Scholar]
- 29.Johnson J R, Russo T A, Scheutz F, Brown J J, Zhang L, Palin K, Rode C, Bloch C, Marrs C F, Foxman B. Discovery of disseminated J96-like strains of uropathogenic Escherichia coli O4:H5 containing genes for both PapGJ96 (“class I”) and PrsGJ96 (“class III”) Gal(α1–4)Gal-binding adhesins. J Infect Dis. 1997;175:983–988. doi: 10.1086/514006. [DOI] [PubMed] [Google Scholar]
- 30.Johnson J R, Russo T A, Tarr P I, Carlino U, Bilge S S, Vary J C J, Stell A L. Molecular epidemiological and phylogenetic associations of two novel putative virulence genes, iha and iroNE. coli, among Escherichia coli isolates from patients with urosepsis. Infect Immun. 2000;68:3040–3047. doi: 10.1128/iai.68.5.3040-3047.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Johnson J R, Stapleton A E, Russo T A, Scheutz F S, Brown J J, Maslow J N. Characteristics and prevalence within serogroup O4 of a “J96-like” clonal group of uropathogenic Escherichia coli O4:H5 containing the “class I” and “class III” alleles of papG. Infect Immun. 1997;65:2153–2159. doi: 10.1128/iai.65.6.2153-2159.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Johnson J R, Stell A, Delavari P. Canine feces as a reservoir of extraintestinal pathogenic Escherichia coli. Infect Immun. 2001;69:1306–1314. doi: 10.1128/IAI.69.3.1306-1314.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Johnson J R, Stell A L. Extended virulence genotypes of Escherichia coli strains from patients with urosepsis in relation to phylogeny and host compromise. J Infect Dis. 2000;181:261–272. doi: 10.1086/315217. [DOI] [PubMed] [Google Scholar]
- 34.Johnson J R, Stell A L, Delavari P, Murray A C, Kuskowski M, Gaastra W. Phylogenetic and pathotypic similarities between Escherichia coli isolates from urinary tract infections in dogs and extraintestinal infections in humans. J Infect Dis. 2001;183:897–906. doi: 10.1086/319263. [DOI] [PubMed] [Google Scholar]
- 35.Johnson J R, Stell A L, Scheutz F, O'Bryan T T, Russo T A, Carlino U B, Fasching C C, Kavle J, van Dijk L, Gaastra W. Analysis of F antigen-specific papA alleles of extraintestinal pathogenic Escherichia coli using a novel multiplex PCR-based assay. Infect Immun. 2000;68:1587–1599. doi: 10.1128/iai.68.3.1587-1599.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kao J-S, Stucker D M, Warren J W, Mobley H L T. Pathogenicity island sequences of pyelonephritogenic Escherichia coli CFT073 are associated with virulent uropathogenic strains. Infect Immun. 1997;65:2812–2820. doi: 10.1128/iai.65.7.2812-2820.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Knapp S, Hacker J, Then I, Muller D, Goebel W. Multiple copies of hemolysin genes and associated sequences in the chromosomes of uropathogenic Escherichia coli strains. J Bacteriol. 1984;159:1027–1033. doi: 10.1128/jb.159.3.1027-1033.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Korhonen T K, Valtonen M V, Parkkinen J, Vaisanen-Rhen V, Fine J, Orskov F, Orskov I, Svenson S B, Makela P H. Serotypes, hemolysin production, and receptor recognition of Escherichia coli strains associated with neonatal sepsis and meningitis. Infect Immun. 1985;48:486–491. doi: 10.1128/iai.48.2.486-491.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Korhonen T K, Virkola R, Vaisanen-Rhen V, Holthofer H. Binding of purified Escherichia coli O75X adhesin to frozen sections of human kidney. FEMS Lett. 1986;35:313–318. [Google Scholar]
- 40.Kumar S, Tamura K, Nei M. MEGA: Molecular Evolutionary Genetics Analysis, version 1.01. University Park, Pa: The Pennsylvania State University; 1993. [Google Scholar]
- 41.Kunin C M, Hua T H, Krishnan C, Van Arsdale White L, Hacker J. Isolation of a nicotinamide-requiring clone of Escherichia coli O18:K1:H7 from women with acute cystitis: resemblance to strains found in neonatal meningitis. Clin Infect Dis. 1993;16:412–416. doi: 10.1093/clind/16.3.412. [DOI] [PubMed] [Google Scholar]
- 42.Langermann S, Mollby R, Burlein J E, Palaszynski S R, Auguste C G, DeFusco A, Strouse R, Schenerman M A, Hultgren S J, Pinkner S, Winberg J, Guldevall L, Soderhall M, Ishikawa K, Normark S, Koenig S. Vaccination with FimH adhesin protects cynomolgus monkeys from colonization and infection by uropathogenic Escherichia coli. J Infect Dis. 2000;181:774–778. doi: 10.1086/315258. [DOI] [PubMed] [Google Scholar]
- 43.Langermann S, Palaszynski S, Barnhart M, Auguste G, Pinkner J S, Burlein J, Barren P, Koenig S, Leath S, Jones C H, Hultgren S J. Prevention of mucosal Escherichia coli infection by FimH-adhesin-based systemic vaccination. Science. 1997;276:607–611. doi: 10.1126/science.276.5312.607. [DOI] [PubMed] [Google Scholar]
- 44.Lawrence J G, Ochman H. Molecular archaeology of the Escherichia coli genome. Evolution. 1998;95:9413–9417. doi: 10.1073/pnas.95.16.9413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lecointre G, Rachdi L, Darlu P, Denamur E. Escherichia coli molecular phylogeny using the incongruence length difference test. Mol Biol Evol. 1998;15:1685–1695. doi: 10.1093/oxfordjournals.molbev.a025895. [DOI] [PubMed] [Google Scholar]
- 46.Low D, David V, Lark D, Schoolnik G, Falkow S. Gene clusters governing the production of hemolysin and mannose-resistant hemagglutination are closely linked in Escherichia coli serotype O4 and O6 isolates from urinary tract infection. Infect Immun. 1984;43:353–358. doi: 10.1128/iai.43.1.353-358.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Lund B, Lindberg F, Marklund B-I, Normark S. Tip proteins of pili associated with pyelonephritis: new candidates for vaccine development. Vaccine. 1988;6:110–112. doi: 10.1016/s0264-410x(88)80010-0. [DOI] [PubMed] [Google Scholar]
- 48.Maiden C J, Bygraves J A, Feil E, Morelli G, Russwell J E, Urwin R, Zhang Q, Zhou J, Zurth K, Caugant D A, Feavers I M, Achtman M, Spratt B G. Multilocus sequence typing: a portable approach to the identification of clones within populations of pathogenic microorganisms. Proc Natl Acad Sci USA. 1998;95:3140–3145. doi: 10.1073/pnas.95.6.3140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Maslow J N, Mulligan M E, Adams K S, Justis J C, Arbeit R D. Bacterial adhesins and host factors: role in the development and outcome of Escherichia coli bacteremia. Clin Infect Dis. 1993;17:89–97. doi: 10.1093/clinids/17.1.89. [DOI] [PubMed] [Google Scholar]
- 50.Maslow J N, Whittam T S, Gilks C F, Wilson R A, Mulligan M E, Adams K S, Arbeit R D. Clonal relationships among bloodstream isolates of Escherichia coli. Infect Immun. 1995;63:2409–2417. doi: 10.1128/iai.63.7.2409-2417.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Milkman R, Bridges M M. Molecular evolution of the Escherichia coli chromosome. III. Clonal frames. Genetics. 1990;126:505–517. doi: 10.1093/genetics/126.3.505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Mobley H L T, Jarvis K G, Elwood J P, Whittle D I, Lockatell C V, Russell R G, Johnson D E, Donnenberg M S, Warren J W. Isogenic P-fimbrial deletion mutants of pyelonephritogenic Escherichia coli: the role of αGal(1–4)-βGal binding in virulence of a wild-type strain. Mol Microbiol. 1993;10:143–155. doi: 10.1111/j.1365-2958.1993.tb00911.x. [DOI] [PubMed] [Google Scholar]
- 53.Ochman H, Selander R K. Evidence for clonal population structure in Escherichia coli. Proc Natl Acad Sci USA. 1984;81:198–201. doi: 10.1073/pnas.81.1.198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Ochman H, Selander R K. Standard reference strains of Escherichia coli from natural populations. J Bacteriol. 1984;157:690–693. doi: 10.1128/jb.157.2.690-693.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.O'Hanley P. Prospects for urinary tract infection vaccines. In: Mobley H L T, Warren J W, editors. Urinary tract infections: molecular pathogenesis and clinical management. Washington, D.C.: ASM Press; 1996. pp. 405–425. [Google Scholar]
- 56.Olesen B, Kolmos H J, Orskov F, Orskov I. A comparative study of nosocomial and community-acquired strains of Escherichia coli causing bacteraemia in a Danish university hospital. J Hosp Infect. 1995;31:295–305. doi: 10.1016/0195-6701(95)90208-2. [DOI] [PubMed] [Google Scholar]
- 57.Orskov I, Orskov F. Escherichia coli in extra-intestinal infections. J Hyg (London) 1985;95:551–575. doi: 10.1017/s0022172400060678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Phillips I, Eykyn S, King A, Grandsden W R, Rowe B, Frost J A, Gross R J. Epidemic multiresistant Escherichia coli infection in West Lambeth health district. Lancet. 1988;i:1038–1041. doi: 10.1016/s0140-6736(88)91853-3. [DOI] [PubMed] [Google Scholar]
- 59.Picard B, Journet-Mancy C, Picard-Pasquier N, Goullet P. Genetic structures of the B2 and B1 Escherichia coli strains responsible for extra-intestinal infections. J Gen Microbiol. 1993;139:3079–3088. doi: 10.1099/00221287-139-12-3079. [DOI] [PubMed] [Google Scholar]
- 60.Picard B, Sevali Garcia J, Gouriou S, Duriez P, Brahimi N, Bingen E, Elion J, Denamur E. The link between phylogeny and virulence in Escherichia coli extraintestinal infection. Infect Immun. 1999;67:546–553. doi: 10.1128/iai.67.2.546-553.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Plos K, Hull S I, Hull R A, Levin B R, Orskov I, Orskov F, Svanborg-Edén C. Distribution of the p-associated-pilus (pap) region among Escherichia coli from natural sources: evidence for horizontal gene transfer. Infect Immun. 1989;57:1604–1611. doi: 10.1128/iai.57.5.1604-1611.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Prats G, Navarro F, Mirelis B, Dalmau D, Margall N, Coll P, Stell A, Johnson J R. Escherichia coli serotype O15:K52:H1 as a uropathogenic clone. J Clin Microbiol. 2000;38:201–209. doi: 10.1128/jcm.38.1.201-209.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Reid S D, Herbelin C J, Bumbaugh A C, Selander R K, Whittam T S. Parallel evolution of virulence in pathogenic Escherichia coli. Nature. 2000;406:64–67. doi: 10.1038/35017546. [DOI] [PubMed] [Google Scholar]
- 64.Russo T A, Carlino U B, Mong A, Jodush S T. Identification of genes in an extraintestinal isolate of Escherichia coli with increased expression after exposure to human urine. Infect Immun. 1999;67:5306–5614. doi: 10.1128/iai.67.10.5306-5314.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Russo T A, Johnson J R. A proposal for an inclusive designation for extraintestinal pathogenic Escherichia coli: ExPEC. J Infect Dis. 2000;181:1753–1754. doi: 10.1086/315418. [DOI] [PubMed] [Google Scholar]
- 66.Saitou N, Nei M. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol. 1987;4:406–425. doi: 10.1093/oxfordjournals.molbev.a040454. [DOI] [PubMed] [Google Scholar]
- 67.Schubert S, Rakin A, Karch H, Carniel E, Heesemann J. Prevalence of the “high-pathogenicity island” of Yersinia species among Escherichia coli strains that are pathogenic to humans. Infect Immun. 1998;66:480–485. doi: 10.1128/iai.66.2.480-485.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Selander R K, Caugant D A, Ochman H, Musser J M, Gilmour M N, Whittam T S. Methods of multilocus enzyme electrophoresis for bacterial population genetics and systematics. Appl Environ Microbiol. 1986;51:873–884. doi: 10.1128/aem.51.5.873-884.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Selander R K, Korhonen T K, Väisänen-Rhen V, Williams P H, Pattison P E, Caugant D A. Genetic relationships and clonal structure of strains of Escherichia coli causing neonatal septicemia and meningitis. Infect Immun. 1986;52:213–222. doi: 10.1128/iai.52.1.213-222.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Siitonen A. What makes Escherichia coli pathogenic? Ann Med. 1994;26:229–231. doi: 10.3109/07853899409147895. [DOI] [PubMed] [Google Scholar]
- 71.Sokal R R, Sneath P H A. Principles of numerical taxonomy. W. H. San Francisco, Calif: Freeman; 1963. [Google Scholar]
- 72.Stapleton A, Stamm W E. Prevention of urinary tract infection. Infect Dis Clin N Am. 1997;11:719–733. doi: 10.1016/s0891-5520(05)70382-2. [DOI] [PubMed] [Google Scholar]
- 73.Swenson D L, Bukanov N O, Berg D E, Welch R A. Two pathogenicity islands in uropathogenic Escherichia coli J96: cosmid cloning and sample sequencing. Infect Immun. 1996;64:3736–3743. doi: 10.1128/iai.64.9.3736-3743.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Tarr P I, Bilge S S, Vary J C, Jr, Jelacic S, Habeeb R L, Ward T R, Baylor M R, Besser T E. Iha: a novel Escherichia coli O157:H7 adherence-conferring molecule encoded on a chromosomal region of conserved structure. Infect Immun. 2000;68:1400–1407. doi: 10.1128/iai.68.3.1400-1407.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Thompson J D, Higgins D G, Gibson T J. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Vaisanen-Rhen V, Elo J, Vaisanen E, Siitonen A, Orskov I, Orskov F, Svenson S B, Makela P H, Korhonen T. P-fimbriated clones among uropathogenic Escherichia coli strains. Infect Immun. 1984;43:149–155. doi: 10.1128/iai.43.1.149-155.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Wang G, Whittam T S, Berg C M, Berg D E. RAPD (arbitrary primer) PCR is more sensitive than multilocus enzyme electrophoresis for distinguishing related bacterial strains. Nucleic Acids Res. 1993;21:5930–5933. doi: 10.1093/nar/21.25.5930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Whittam T A, Wilson R A. Genetic relationships among pathogenic Escherichia coli of serogroup O157. Infect Immun. 1988;56:2467–2473. doi: 10.1128/iai.56.9.2467-2473.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Whittam T S, Ochman H, Selander R K. Multilocus genetic structure in natural populations of Escherichia coli. Popul Biol. 1983;80:1751–1755. doi: 10.1073/pnas.80.6.1751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Whittam T S, Wolfe M L, Wilson R A. Genetic relationships among Escherichia coli isolates causing urinary tract infections in humans and animals. Epidemiol Infect. 1989;102:37–46. doi: 10.1017/s0950268800029666. [DOI] [PMC free article] [PubMed] [Google Scholar]