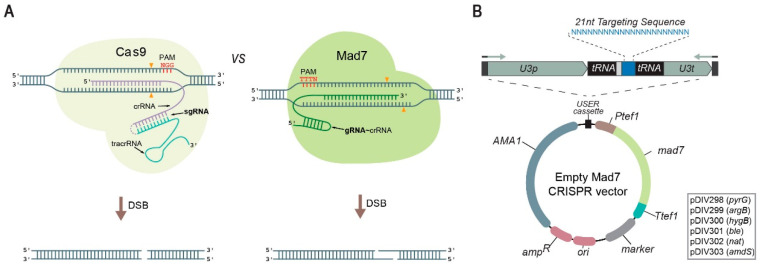
Figure 1.
A Mad7 system for genetic engineering of fungi. (A) Schematic representations of Cas9 (left) and Mad7 (right) CRISPR nucleases. Scissile bonds in the target sequences are marked with orange triangles [24,25]. The positions of PAM sequences, as well as the orientation and the pairing of the sgRNA of Cas9 (fusion of tracrRNA and crRNA is indicated by the dotted line) and the gRNA of Mad7 (note that Mad7 has a natural single gRNA(=crRNA)) to the target template, are shown. (B) Mad7 vectors available for fungal genetic engineering. The vectors are shuttle vectors that can propagate in E. coli using ampicillin selection and in fungi using the AMA1 sequence and one of the selection markers indicated in the box. All vectors contain a mad7 gene and a USER cassette for insertion of the gRNA encoding gene by e.g., USER fusion of two PCR fragments, see Figure S1. Expression of the gRNA gene results in a composite transcript where the gRNA is released by the endogenous tRNA maturation machinery; see text for details.