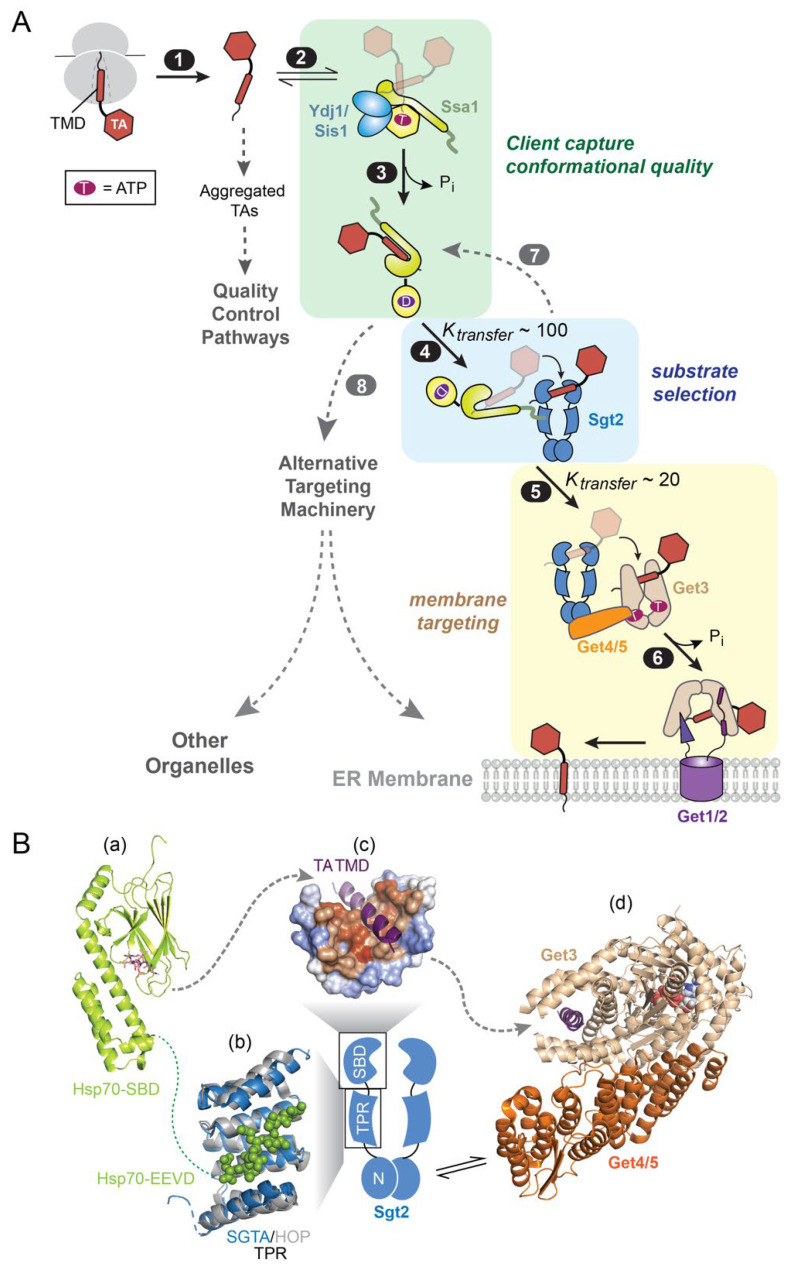
Figure 4.
An Hsp70-cochaperone cascade mediates TA targeting to the ER via the GET pathway. (A) Scheme of the molecular steps in the GET pathway. TAs released from the ribosome (step 1) associate with the major cytosolic Hsp70 in yeast, Ssa1 (step 2). Two JDPs, Ydj1 and Sis1, catalyze ATP hydrolysis on Ssa1 and assist in substrate trapping (step 3). TA-bound Ssa1 associates with the cochaperone Sgt2 via its C-terminal EEVD motif, forming the first transfer complex in which TA is loaded onto Sgt2 (step 4). Get4/5 bridges between Sgt2 and the targeting factor Get3, forming the second transfer complex in which TA is relayed onto Get3 (step 5). TA-activated ATP hydrolysis on Get3 drives its dissociation from Get4/5 and association with the Get1/2 receptor complex at the ER membrane (step 6), via which the TA substrate is inserted. The dashed lines indicate that suboptimal TA substrates can be rejected by Sgt2 (step 7) and re-routed to alternative targeting pathways (step 8). (B) Summary of available structural information on the TA transfer complexes in the GET pathway. Panel (a), structure of the DnaK SBD bound to the NRLLLTG peptide (shown as stick; PDB 1dkz). Panel (b), the structure of the TPR domain of SGTA (blue; PDB 5lyn) is overlayed onto that of the TPR1 domain of HOP (grey) bound to the HSC70 C-terminal peptide GPTIEEVD (shown in spacefill; PDB 1elw). Panel (c), molecular model of the Sgt2/SGTA SBD bound to a TA-TMD (purple), shown in surface representation with hydrophobic residues in orange and hydrophilic residues in blue. Adapted from [73]. Panel (d), the structure of Get4/5 (orange) bound to Get3 (tan; PDB 4pwx) is overlayed onto that of Get3 bound to the TMD of the model TA Pep12 (purple; PDB 4xtr). The dotted line indicates unresolved flexible regions, and dashed arrows depict paths of TA transfer.