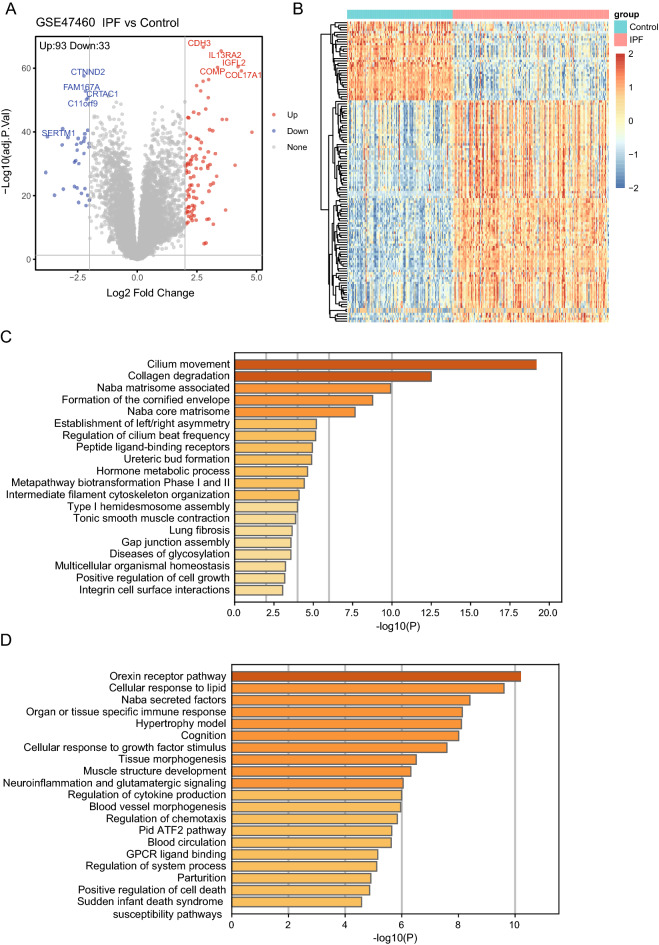
Figure 2.
Differential gene expression analysis in IPF. (A) Volcano plot of differential expression analysis results. The abscissa is log2Fold Change and the ordinate is –log10 (adj.P value). The upper right part has a adj.P value less than 0.05 and a fold change greater than 2, indicating significant DEGs with higher expression levels. The upper left part has a adj.P value less than 0.05 and a fold change less than − 2, indicating significant DEGs with reduced expression. The gray dots represent the remaining stable genes. (B) Heatmap of DEGs. The colors in the graph from red to blue indicate high to low expression. On the upper part of the heatmap, the red band indicates the disease samples and the blue band indicates the normal samples. C-D. Matescape toll function enrichment results bar graph. The x-axis represents −log10(adj P) values and the y-axis represents enriched pathways. Pathways with Log10(P value) of > 2.5 are marked and shown in the figure. (C) shows a bar graph of the enriched pathways that were significantly up-regulated in IPF patients compared to healthy controls. (D) shows a bar graph of the enrichment pathway results that were significantly downregulated in IPF patients compared to healthy controls.