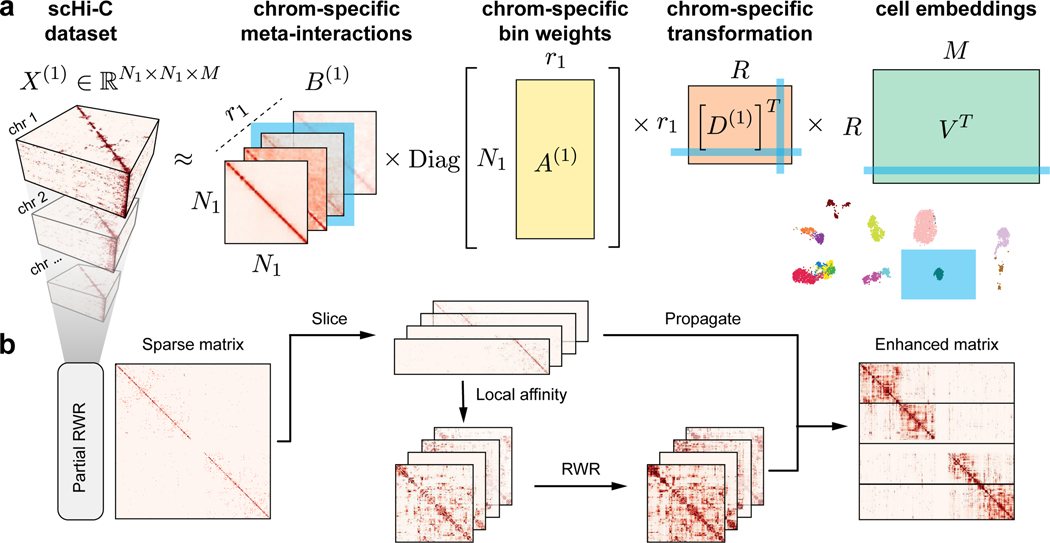
Figure 1: Overview of Fast-Higashi.
a. Workflow of the Fast-Higashi algorithm. Given an input scHi-C dataset of k chromosomes, Fast-Higashi models it as k 3-way tensors. The tensor of chromosome c is denoted by X(c), where the first two dimensions correspond to genomic bins and the last dimension corresponds to the single cells. Fast-Higashi then decomposes the tensors X(c) into four factors: a set of meta-interactions (B(c)), a genomic bin weights indicating importance for each bin (A(c)), a cell embedding matrix V that is shared across all chromosomes, and a chromosome-specific transformation matrix D(c) that transforms the shared cell embeddings into chromosome-specific ones. b. Workflow of the partial random walk with restart (Partial RWR) algorithm. The Partial RWR is integrated into the Fast-Higashi framework. When calculating the decomposed factors for frontal slices of the tensor X(c), the corresponding slices would be imputed through Partial RWR first. The imputation process includes the calculation of local affinity, standard RWR algorithm, and information propagation using both sliced tensor and the RWR imputed affinity matrix.