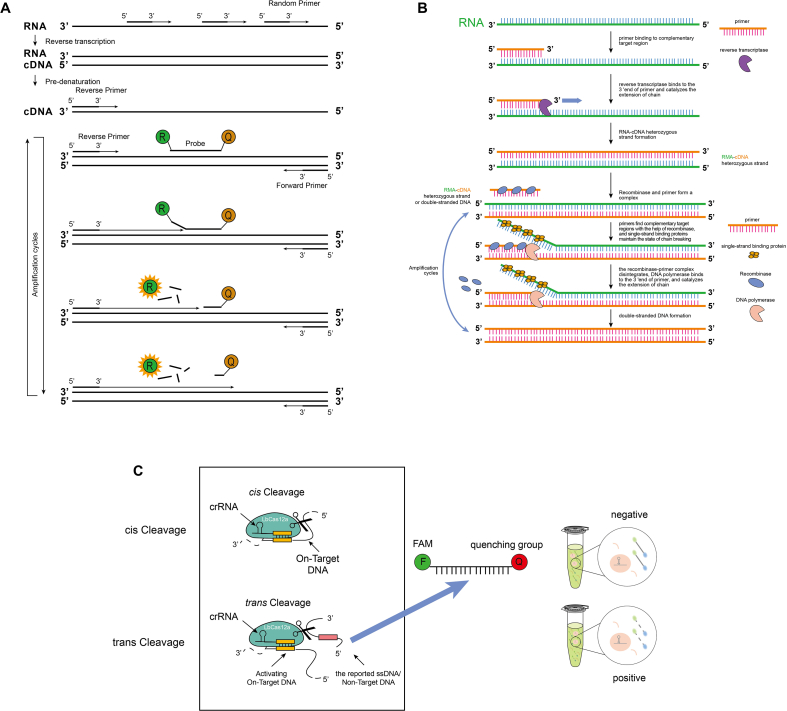
Fig. 1.
Schematics showing the principles of RT-PCR, RPA, and CRISPR/Cas12a detection systems. The commercial SARS-CoV-2 kit (BioGerm, Shanghai, China) is designed based on RPA and CRISPR. (A) RT-PCR: N gene target was amplified and evaluated using real-time Taqman PCR. (B) RPA: At 42 °C, RNA was first reverse transcribed to cDNA using reverse transcriptase. Recombinase proteins form complexes with each primer, which scan cDNA for homologous sequences. DNA polymerase uses designed primers to transcribe the complementary strand of cDNA and forms a double-stranded DNA (dsDNA) template. The recombinase/primer complex binds to dsDNA and promotes strand displacement by the primer at the cognate site, and single stranded binding proteins stabilize the displaced DNA chain. The recombinase disassembles and a strand displacing DNA polymerase binds to the 3′-end of the primers, which elongates the primer. The exponential amplification is then achieved by cyclic repetition of this process. (C) Detector assay: The target sequence is amplified by the RPA reaction, followed by mixing with the CRISPR/LbCas12a reagents for cleavage. LbCas12a utilizes the crRNA as guides to bind cognate DNA sequences. LbCas12a is then activated for on-target DNA cleavage (cis cleavage) and non-target single-stranded DNA cleavage (trans/collateral cleavage). The reported signal-stranded DNA (ssDNA) includes a fluorophore-quencher pair, and the cleavage events will free the fluorophore from its quencher, in effect activating fluorescence that can be measured.