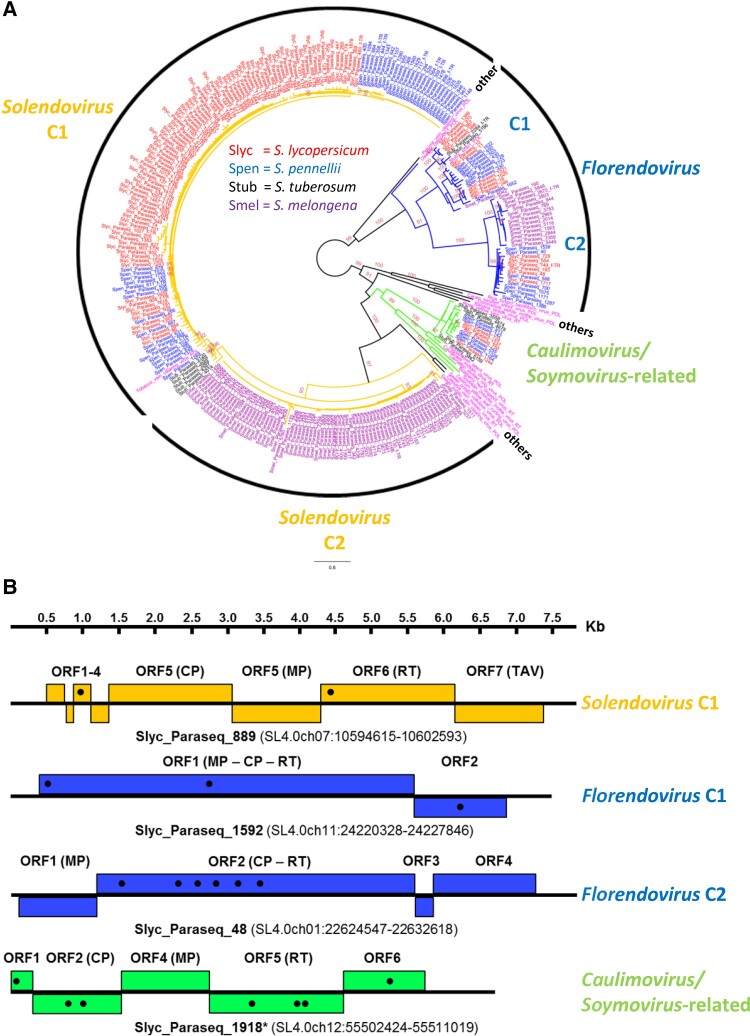
Fig. 1.
Summarized overview of endogenized pararetroviral sequences in exemplary Solanum genomes. (A) Unrooted phylogenetic relationship of recognized pararetroviral-related reverse-transcriptases (RTs) protein sequences with >300 amino-acids (as a conservative threshold to ensure robust confidence in alignment) called within current Solanum lycopersicum (red), Solanum pennellii (blue), Solanum tuberosum (black), and Solanum melongena (violet) genomes. Poly-protein (POL) or RT sequences from 16 different pararetroviruses, along with two complete reported endogenized elements from Florendovirus (StubV_scSt1 and OsatBV_compAsc1 from S. tuberosum and Oryza sativa, respectively), were added in pink to expose phylo-group relatedness. Shown ≥ 90 bootstrap support was considered significant. Phyletic relationships are highlighted for Solendovirus (orange; sub-clades C1 and C2, but note that Sweet potato vein clearing virus would represent an additional sub-clade), Florendovirus (blue; sub-clades C1 and C2) and Caulimovirus/Soymovirus-related group (green). Black phyletic lines represent other/s (Badnavirus, Tungrovirus, Cavemovirus, Petuvirus and Rosadnavirus). f_TR denotes sequences bearing recognized flanking tandem-repeats. (B) Schematic representation of exemplary S. lycopersicum non-truncated EPRVs assembled from distinct phylo-groups. Boxes represent ORFs, with black dots indicating positions with premature stop codons, manually curated using as template other EPRV relatives of the corresponding phylogenetic clade. ORFs encoding conserved domains are indicated between brackets (CP, capsid protein; MP, movement protein; RT, reverse-transcriptase; TAV, transactivator/viroplasmin; ORFX, unknown). Chromosomal coordinates of each non-truncated EPRV are indicated. Note that Slyc_Paraseq_1918* is a portion of an original parasequence which presented a rearrangement at the 5′ end, additionally extending the otherwise complete non-truncated element here presented.