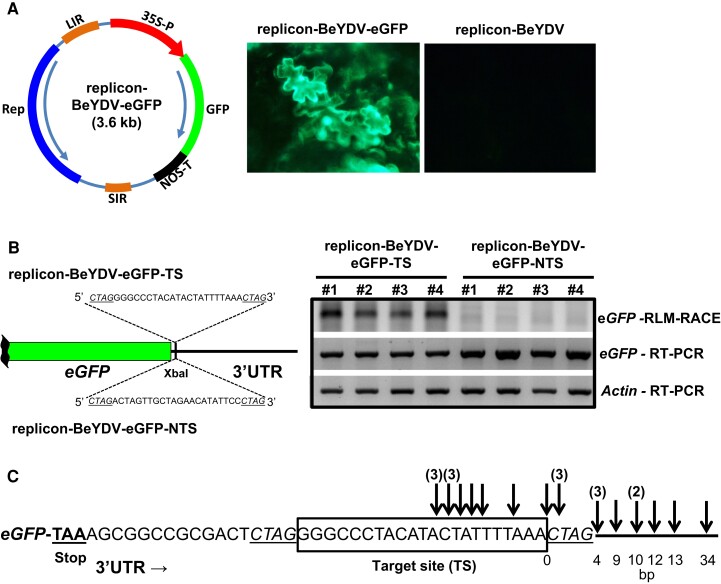
Fig. 4.
Exemplary target-site for private siRNAs mapping TSA10 promotes cleavage of a viral replicon-derived reporter transcript. (A) Schematic representation of the self-replicating artificial BeYD-based viral replicon generated after infiltration of Solanum lycopersucum cotyledons with Agrobacterium tumefaciens strain carrying a pRIC3.0-eGFP-derived plasmid. Microscope pictures of cotyledons under UV light were taken three days post-infiltration and demonstrated eGFP expression in S. lycopersicum cells only when the plasmid carried the reporter (left); an eGFP-less pRIC3.0-derived plasmid was assayed as negative control (right). (B) Left: schematic representation of the manipulated segment of the viral replicon in which a short target-site (TS) for TSA10-derived siRNAs and a corresponding non-target-site (NTS, negative control) were introduced. Right: agarose gels are shown for RLM-RACE and RT-PCR from cotyledon total RNA sampled three days post-infiltration in independent S. lycopersicum plants. Upper gel, eGFP mRNA cleaved products detected by RLM-RACE with expected size ∼240 bp; middle gel, eGFP RT-PCR; lower gel, Actin RT-PCR (housekeeping gene). (C) Sanger sequencing analysis of 23 independent RLM-RACE products after cloning DNA fragments from the upper gel. Arrows represent exact cleavage position and above numbers indicate clones’ count if beyond one. Distance to the 3′ end of target-site in basepairs.