Figure 5. Striatal genome editing following RNP NC delivery is observed primarily in medium spiny neurons.
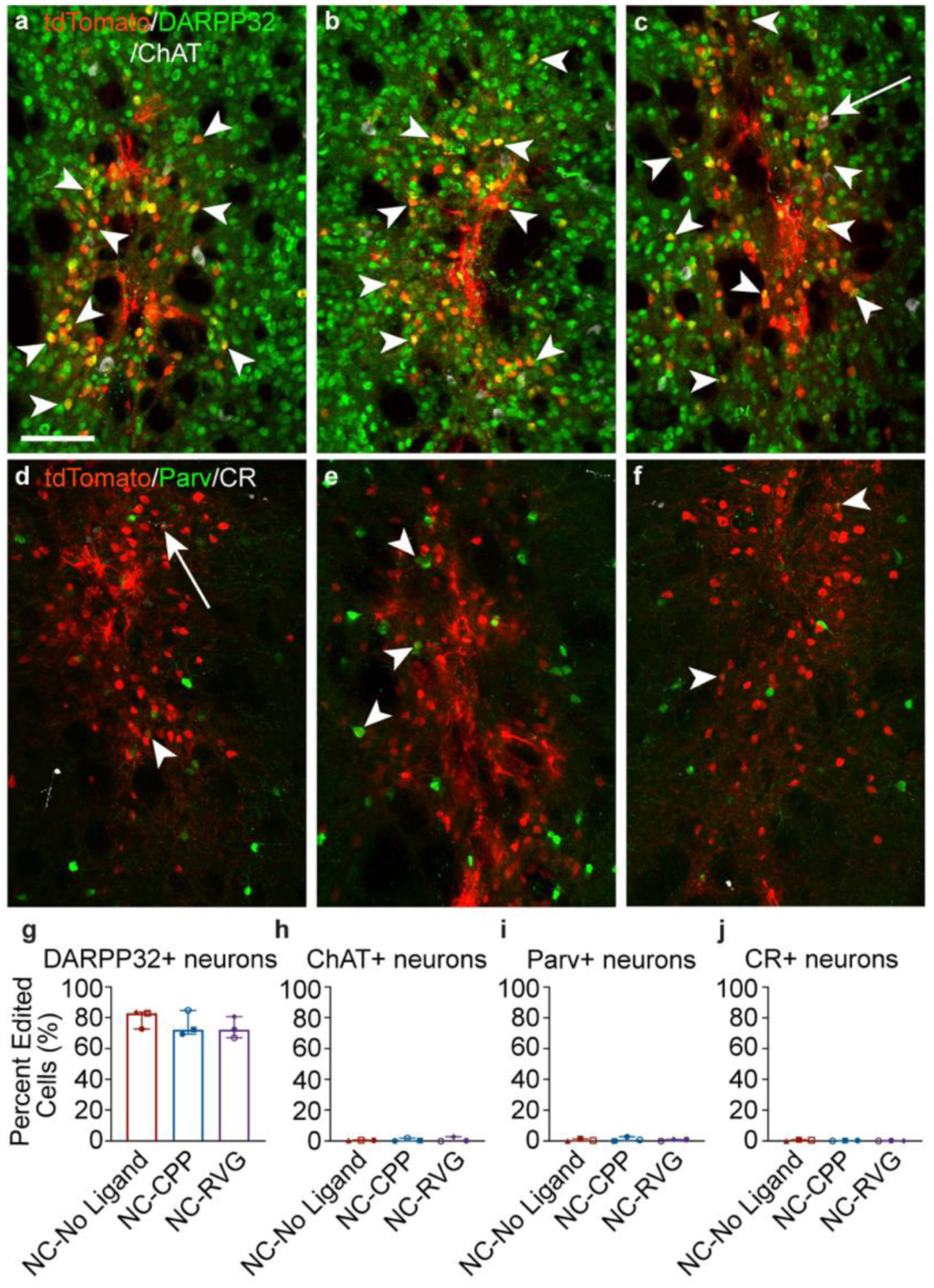
a – c, Genome-edited neurons in the mouse striatum (tdTomato+; red) are primarily medium spiny neurons, as indicated by DARPP32 (green) co-labeling (white arrowheads). A small number of choline acetyl transferase (ChAT)+ (white) neurons are genome-edited (white arrows). This pattern was similar across NC treatment groups (a, NC-No Ligand animal J9; b, NC-CPP animal J17; NC-RVG animal J5; Supp. Table 1). d – f, Genome-edited parvalbumin (Parv+; white arrowheads) and calretinin (CR+; white arrows) neurons were occasionally observed (a, NC-No Ligand animal J8; b, NC-CPP animal J13; NC-RVG animal J4; Supp. Table 1). a – f, scale = 100 μm. Photomicrographs show maximum intensity projection of three focal planes covering 10 μm. Individual channels were adjusted for brightness as needed (Supp. Table 4). Percentage of tdTomato+ neurons that co-labeled for DARPP32 (g), ChAT (h), parvalbumin (i), or calretinin (j) across treatment groups. Graphs show median and interquartile range. Differences between groups were not statistically significant. Each animal is shown with a unique color and symbol (Supp. Table 1). CPP, cell-penetrating peptide; DAPI, 4′,6-diamidino-2-phenylindole; DARPP32, dopamine- and cyclic-AMP-regulated phosphoprotein of molecular weight 32 kDa; NC, nanocapsule; RNP, ribonucleoprotein; RVG, rabies virus glycoprotein.
