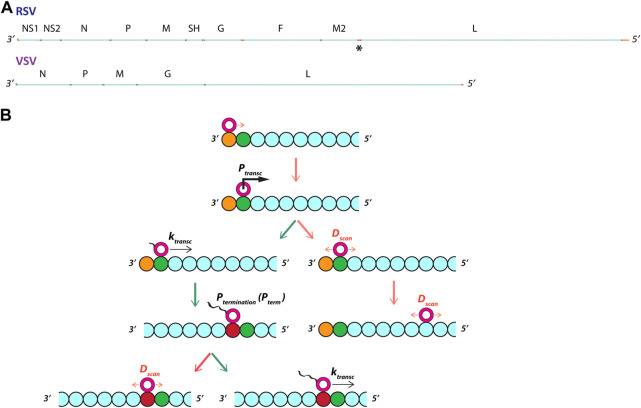
FIGURE 1.
The model: linear respiratory syncytial virus (RSV) and vesicular stomatitis virus (VSV) genomes support the stochastic initiation and termination of transcription by a diffusing viral RNA-dependent RNA polymerase (pol). (A) The genetic structure of RSV and VSV genomes. The modeled RSV genome is 15,222 nt long and contains 10 ORFs with 8 gene junctions and a single short region (68 nt) of overlapping ORFs between genes M2 and L (see black asterisk). The modeled VSV genome is 11,152 nt long and contains 5 ORFs with 4 gene junctions. The genomes were divided into chunks approximating the size of a pol footprint (28 nts). Most of each genome is coding sequence (represented as cyan beads). (B) Essential model phenomena and parameters. A single RNA-dependent RNA polymerase (pol) starts an unbiased random walk at a rate D scan (= 1 genomic chunk per event) at the most 3’ chunk (depicted as a burnt orange bead) of the modeled genome. Transcription initiation occurs with a probability P transc when a pol diffuses onto a genomic chunk containing a gene start (GS) signal (depicted as a green bead). If transcription is not initiated, the unbiased random walk (i.e., diffusion) resumes. If transcription is initiated, the modeled pol state changes and the pol starts translocating 5’ down the genome at a rate k transc (= x genomic chunks per event). Transcription termination occurs with a probability P term when a transcribing pol translocates onto a genomic chunk containing a gene end (GE) signal (depicted as a red bead). If termination occurs, the pol state changes back to non-transcribing and resumes diffusion along the genome at a rate D scan ; if termination does not occur, the pol ‘reads through’ the GE signal and continues transcribing into the next ORF. (Cyan beads represent coding sequence).