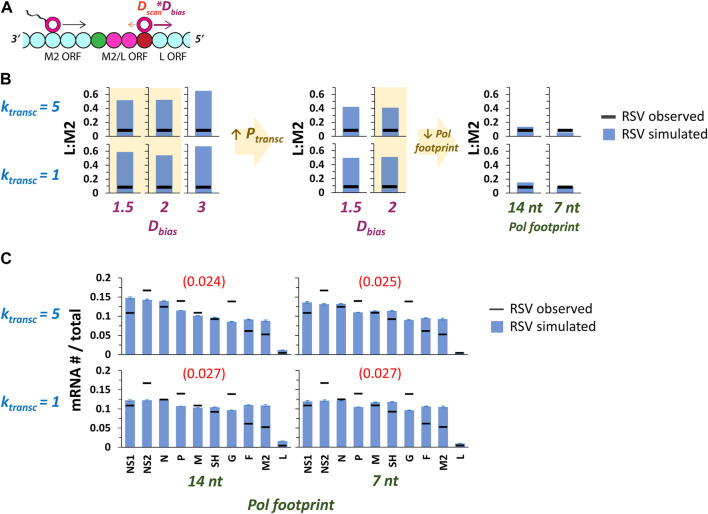
FIGURE 6.
Including a 5’ pol diffusion bias (D bias ) can reproduce the experimentally observed drop in gene expression between RSV genes M2 and L. (A) D bias is expected to have its largest effect on ratios of L mRNA to M2 mRNA levels (L:M2). M2 and L ORFs are depicted as cyan beads (bead size reflects a pol footprint size of 28 nt); the M2/L overlap is depicted as two magenta beads; the L GS signal is depicted as a green bead; the M2 GE signal is depicted as a red bead. (B) A scan of parameter values shows that moderate D bias with reduced pol footprint size can reproduce the experimentally observed value of L:M2 (Rajan et al., 2022). Left panel: histograms show simulated (blue bar) and experimentally observed (horizontal black bar) L:M2 values for two different values of k transc x three different values of D bias (max pol # = 5). The two lower values of D bias tested (highlighted in pale yellow) result in a slightly greater drop in L:M2 than D bias = 3; these values were used in subsequent simulations. Middle panel: histograms show simulated (blue bar) and experimentally observed (horizontal black bar) L:M2 values for two different values of k transc x two different values of D bias under conditions of increased P transc (= max of 0.9). As predicted, an increased P transc resulted in a further decreased L:M2. Simulations with D bias = 2 (results highlighted in pale yellow) were chosen for subsequent simulations. Right panel: histograms show simulated (blue bar) and experimentally observed (horizontal black bar) L:M2 values for two different values of k transc x two different pol footprint sizes (14 and 7 nt) and D bias = 2. A decreased pol footprint size increases the effective distance between the M2 GE and L GS signals, and results in simulated levels of L:M2 that closely match experimental observations. Each data point is the average of three 100,000 event simulations. (C) Global fits of the RSV gene expression data improve with the introduction of D bias and reduced pol footprint size. Simulations of RSV transcription were performed at two different values of k transc x two different values of pol footprint and D bias = 2. Histograms of mRNA # for each RSV gene divided by the total mRNA # depict results from the simulations (blue bars) and average published experimentally observed values (black horizontal bars) (Rajan et al., 2022). Each data point is the average of three 100,000 event simulations; error bars show the standard deviation. The number in parentheses and red above each histogram is the root-mean-square deviation (RMSD) of the simulated gene expression pattern from the experimental observations.